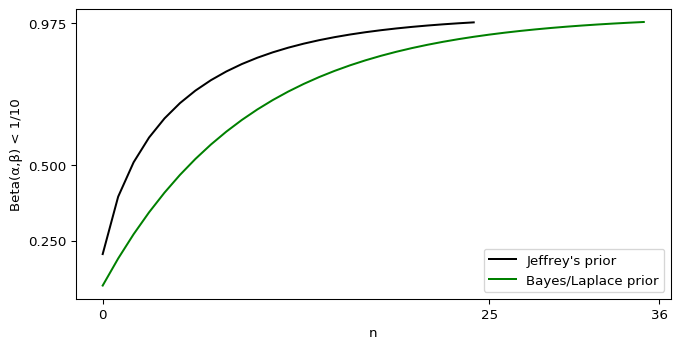
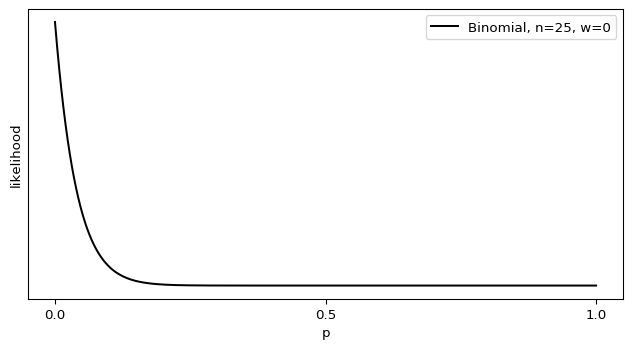
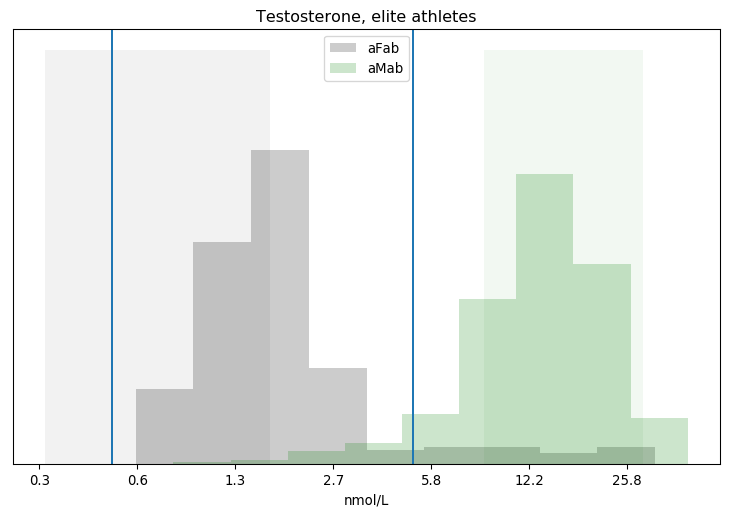
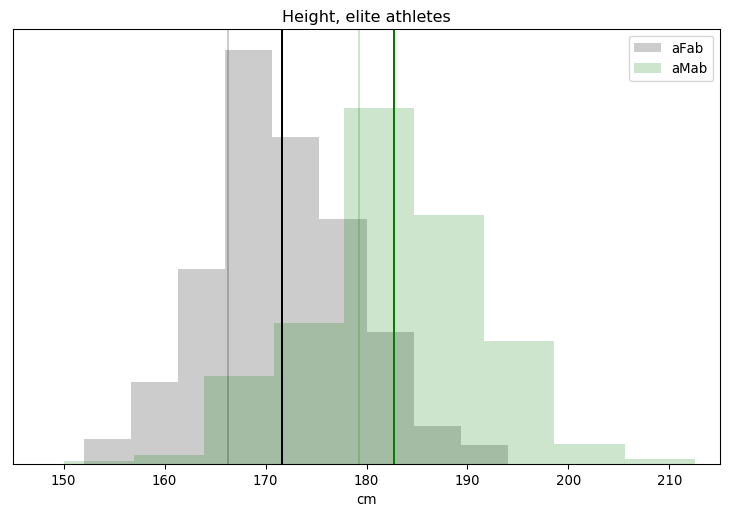
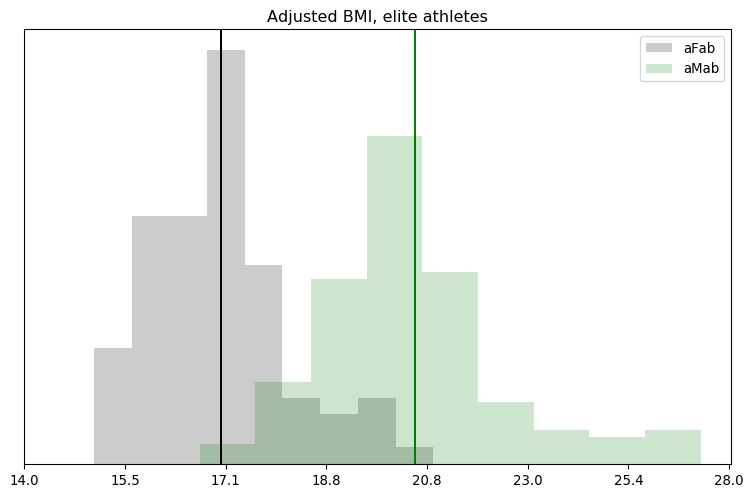
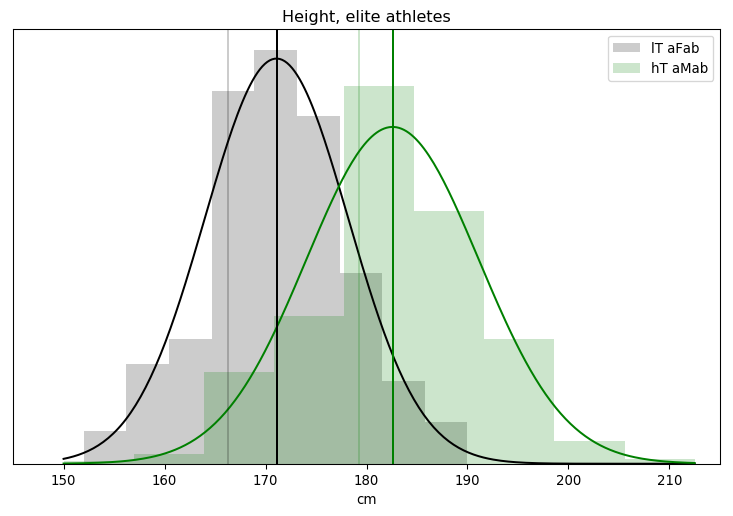
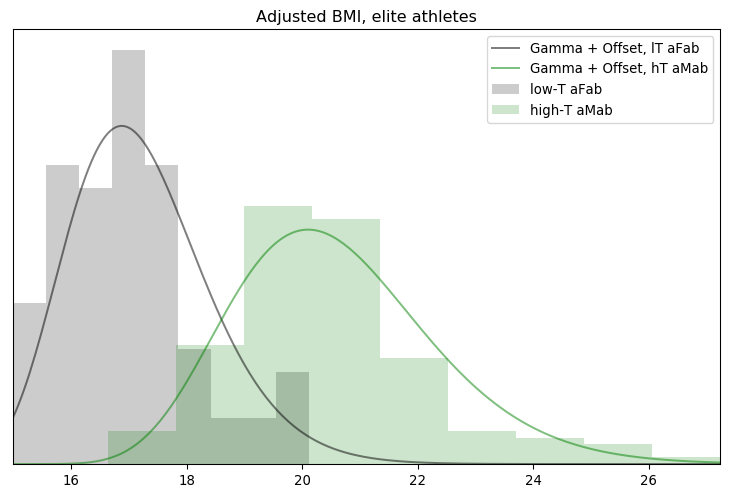
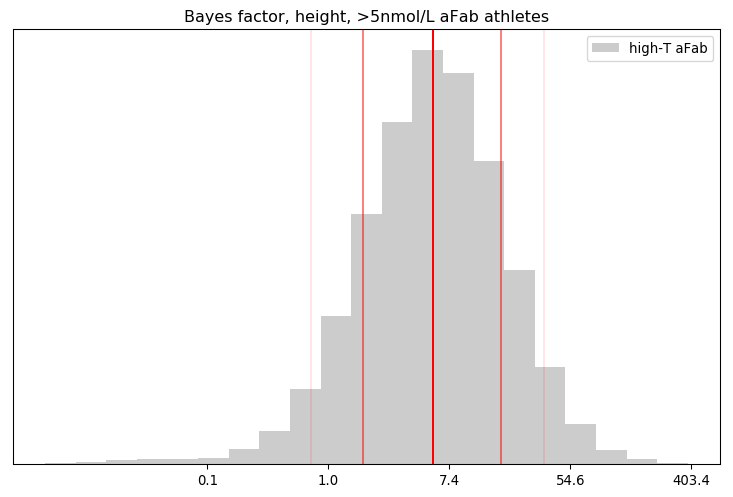
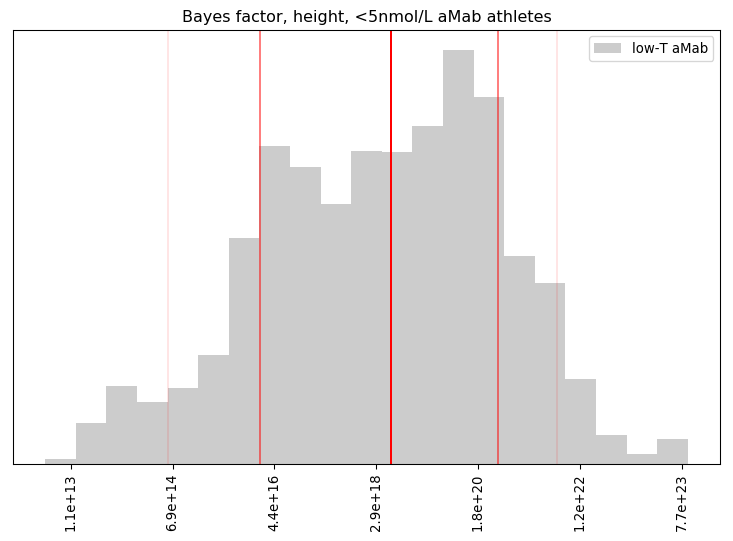
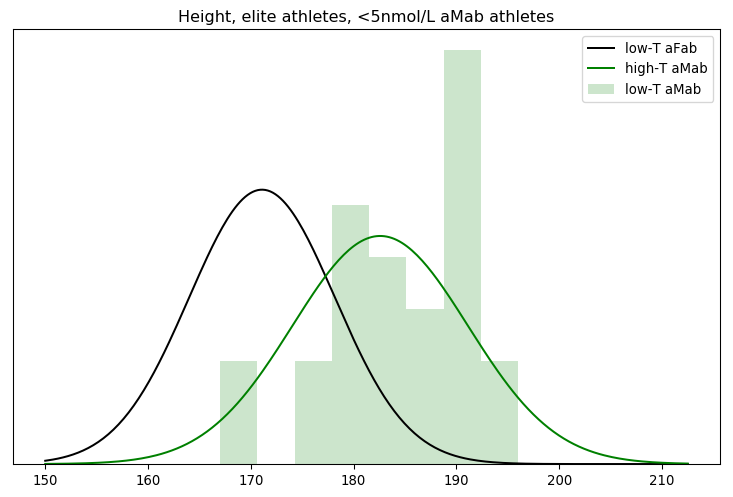
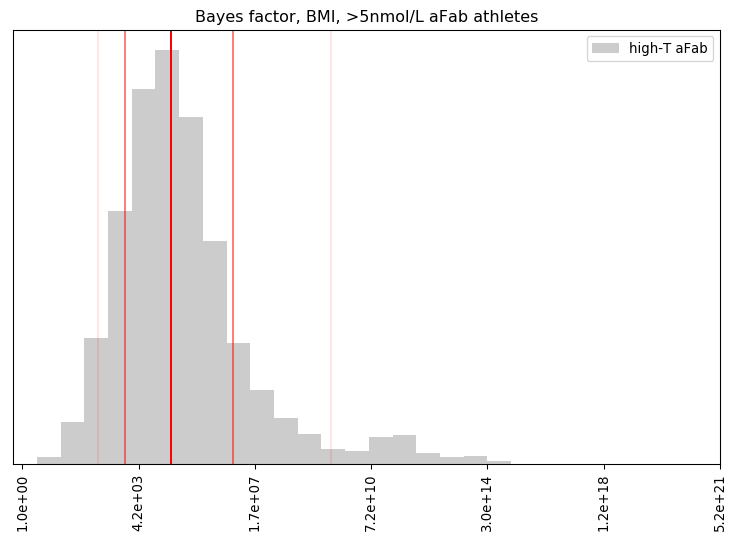
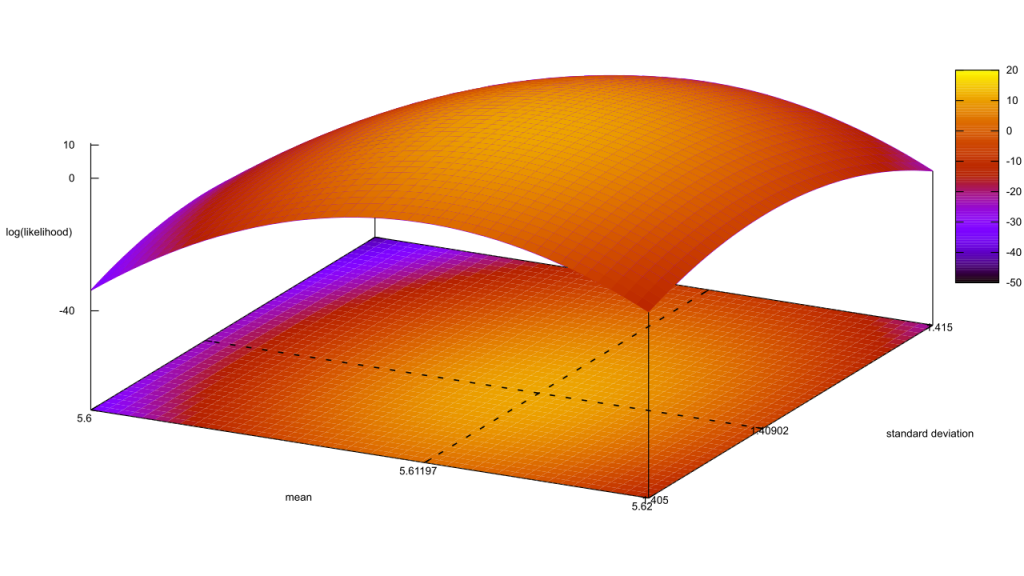
I’ll admit, this fundraiser isn’t exactly twisting my arm. I’ve been mulling over how I’d teach Bayesian statistics for a few years. Overall, I’ve been most impressed with E.T. Jaynes’ approach, which draws inspiration from Cox’s Theorem. You’ll see a lot of similarities between my approach and Jaynes’, though I diverge on a few points. [Read more…]