I can see my future now, for at least the next four months. I have committed myself to record all of my lectures so the students have asynchronous access to the course content to maximize flexibility in case pandemic catastrophe strikes, so what I’ve got to do is:
- Every Monday and Wednesday, record my in-class lecture, then edit it and splice in the Keynote images I use. That means I put in my regular workday, and then when I get home that evening I get to play with Final Cut Pro. Two of those a week means I won’t be doing anything fancy, just dubbing the slides and uploading it.
- Every weekend, I put together a video to describe the lab experiment for the week. The last one might be the longest of the bunch at about a half hour, once the students get into the routine it may not be too bad.
- Every week I also assemble a clutch of problems for the students to solve. Those go on Canvas, our course management system.
There are always glitches. Last week, the audio recording of the lecture was unlistenable, so I had to re-record the whole thing. That was better (but far from perfect) today. Today, though, the in-class technology threw up a whole bunch of problems — nothing worked until I called in IT to fix it, so I lost over 10 minutes to annoying problems. I intensely dislike the way the university has configured the AV in our classrooms.
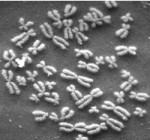
So anyway, here’s today’s lecture. It’s about chromosomes.