Ophelia has summarized a series of science questions Richard Dawkins asked on Twitter. Hey, I thought, I have answers to lots of these — he probably does, too — so I thought I’d address one of them. Maybe I can take a stab at some of the others another time.
I like this one, anyway:
Why do cells have the complete genome instead of just the part that’s needed for their function? Liver cells have muscle-making genes etc.
My short answer: because excising bits of the genome has a high cost and little benefit, and because essentially all of the key exaptations for multicellularity evolved in single-celled organisms, where modifying the DNA archive would have serious consequences for all the daughter cells.
This is an interesting issue, though: different kinds of cells in the same organism express genes that are qualitatively and quantitatively different. Here’s a set of nice graphs in which the relative fraction of different classes of genes in gene transcripts in different cell types were measured. Notice in the list of biological processes that a lot of them, such as the genes involved in transcription and translation and metabolism, are going to be used in all cells, but some, such as neuron-specific or testis-specific genes, are only going to be expressed in some cells.
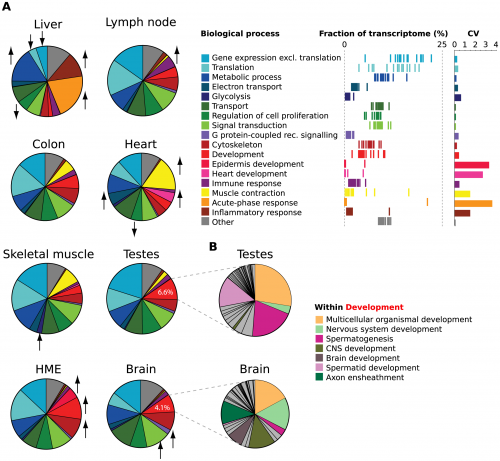
(A) Pie graphs show estimated fraction of cellular transcripts deriving from genes belonging to a set of top-level Gene Ontology Biological Process categories for 7 human tissues and 1 cell line. Fractions were estimated from read density (RPKM) of Ensembl transcripts for each gene. Names of categories, distribution of transcriptome fraction across the samples (each line is a sample), and the coefficients of variation are shown at right. Biological processes with significantly higher or lower densities in individual tissues and cell lines are denoted by arrows. (B) FRACT analysis of sub-categories of the top-level ‘Development’ category in brain and testes.
It also gets complicated because some genes are found in very different forms: there is a kind of universal myosin, myosin I, for instance, that is expressed in all cells as part of the intracellular transport machinery, and then there is a myosin variant, myosin II, that is expressed only as a part of the contractile machinery in muscle. So you might think that it would be more efficient for a skin cell to simply cut out and throw away Myosin II, since it’ll never use it, and keep Myosin I.
But how does the cell determine which genes it will never use? Where does it draw the line? All those testis development genes, for example — I never used many of them until I hit puberty. Wouldn’t it have been terrible if my young toddler testicles threw out a set of unused genes, and then a dozen years later discovered that they had a use, after all? There are a great many genes regulated by timing and signals, and as can be seen in that figure above, every cell has a different expression profile. There are a variety of cells in my skin that are busy replicating and making keratin proteins as a matter of course, but they only switch on cellular repair mechanisms if I cut myself. There are also many genes that get reused in complicated ways, too: the gene even-skipped is first switched on as part of the segment forming process in flies, but it later is switched on again in making neuroblasts, and later still is expressed in axons during pathfinding. Cells would rather recycle genes than throw them away.
These properties are not unique to us mammals, either. Bacteria regulate which genes are turned off and on, too — they change their biochemical behavior in response to signals in their environment. The ability to switch on and switch off genes, without eliminating the DNA, is a solved problem. Life figured that one out a few billion years ago. Key molecules required for multicellular patterns of gene expression first evolved in bacteria — they worked out how to have a cell with the same genetic material behave differently in different circumstances. We came about ready made with a toolkit equipped to have one set of genes turned on in livers, and a different set turned on in muscles, easy.
But, you might think, wouldn’t it be so much more cost-efficient if cells in multicellular organisms just got rid of genes they’d never turn on in their lifetime, once they’ve committed to a certain tissue type? Muscle cells will never make sperm recognition proteins, and liver cells won’t ever have to lift weights, and you could probably cut the amount of DNA in differentiated cells in half with no effect on function.
But that’s penny-wise accounting. In bacteria, only about 2% of the cell’s energy budget is invested in replication — so removing a bit of DNA here and there is only going to shave a tiny amount off the cost of cell division. On the other hand, an amazing 75% is spent on transcribing and translating genes, so efficient mechanisms of simply turning off unused genes reaps huge savings for the cell. Evolving a complex process to pare away unused DNA in terminally differentiated cells simply does not make sense energetically, while simply taking advantage of an already fully implemented and refined process for regulating gene expression…heck, that’s what evolution does best, reusing what’s already there.
By the way, not all cells carry the complete genome: there are also a few cases where the DNA of an organism is modified — the CRISPR system in bacteria, and the somatic recombination system used in vertebrates to generate diverse immunoglobulins. In both of those cases, though, it’s not a mechanism to cut away unused DNA. It’s a specialized process to create variation during an organism’s lifetime to cope with environmental challenges.
Lane N, Martin W. (2010) The energetics of genome complexity. Nature 467(7318):929-34.
Ramsköld D1, Wang ET, Burge CB, Sandberg R (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5(12):e1000598. doi: 10.1371/journal.pcbi.1000598.


Where do red blood cells fit in this economic model?
And platelets!
They’ve completely cut out transcription, so they’ve saved quite a bit of energy there; but the cost is no maintenance and a short life.
I thought I remembered an exception: in some organisms, somatic cell lines lose chromosomes, while the germ cell lines retain all chromosomes.
Experiments on chromosome elimination in the gall midge, Mayetiola destructor
“Cleavage in Cecidomyidae (Diptera) is characterized by the elimination of chromosomes from presumptive somatic nuclei. The full chromosome complement is kept by the germ-line nuclei.
The course of cleavage in Mayetiola destructor (Say) is described. After the fourth division two nuclei lie in the posterior polar-plasm and become associated with polar granules, and fourteen nuclei lie in the rest of the cytoplasm. All the nuclei possess about forty chromosomes. During the fifth division the posterior nuclei do not divide and the polar-plasm becomes constricted to form primordial germ cells (pole cells). The remaining fourteen nuclei divide and lose about thirty-two chromosomes so that twenty-eight nuclei are formed containing only eight chromosomes. These are the presumptive somatic nuclei. During subsequent divisions the pole cell nuclei retain the full chromosome number; these divisions occur less frequently than those of the somatic nuclei.”
Well, that was interesting. And I know absolutely nothing about any of this stuff. When I was in school (early ’70s), I’m pretty sure that at least some of this stuff hadn’t been figured out yet.
You must be a really good teacher PZ. If it wasn’t so damn cold in MN, I’d seriously consider taking some of your classes just to learn something new. But I promised the wife that we would retire someplace warmer so I’m stuck in a freakin red state – the one responsible for both Rand and Mitch.
OK, but how is short life defined as a “cost” in this model?
seems to me, given the role platelets play, it’s actually a net benefit.
something I’m not seeing here?
I’m all for applying cost/benefit analysis in examining trait development, but still unclear on what makes something a cost/benefit here.
…btw, I’m now starting to recall that a discussion of exactly this kind of cost/benefit analysis was something I once participated in as a seminar series when I was a grad student, where we were examining at what level selection acts at (gene/cell/individual/group).
so I’m guessing my current intrigue was likely sparked by that old debate.
TOTALLY OT, I apologize, but PZ: Are you aware of the Octopus Overload extravaganza going on at shirt.woot?!
http://shirt.woot.com/plus/octopus-overload
^^put that in the Lounge.
How much of the above is a consequence of path, i.e evidence for evolution? I can imagine a life-form with a central repository of genetic material, with tissue being constructed and sent to their respective places, perhaps without even any cell structures at all, but I can’t see how to get there without building them from whole cloth.
I forgot about the Lounge. I’m totally smart today. (Sorry, I took a pain pill for my hip *smacks forehead*)
But they are densely packed with with a few mRNAs that have already been transcribed in megakaryocytes. E.g., Sparc/osteonectin mRNA.
It’s not true in many organisms – for example, lampreys shed about 13% of their germline DNA. Jeremiah Smith has papers, Wang and Davis a review, and Jerry Coyne a blog post about it:
Smith, J.J., Baker, C., Eichler, E.E., and Amemiya, C.T. (2012). Genetic consequences of programmed genome rearrangement. Curr. Biol. 22, 1524–1529.
Programmed DNA elimination in multicellular organisms. J Wang, RE Davis – Current opinion in genetics & development, 2014.
http://whyevolutionistrue.wordpress.com/2012/08/26/lampreys-deep-six-their-genomes-during-development/
It should be noted also that a fair number of terminally differentiated cells will do not divide again. Such cells would energetically speaking save bupkiss from excising DNA (and indeed would take a net loss of energy to manufacture the necessary splicing enzymes….)
I always looked at it from an editing perspective. Excising information is harder than adding it, as it takes more choices, and there isn’t anything in a cell to do the deciding.
Now, if DNA was granular, rather than being one long string, it could be done more easily . . .
I am reminded of a creationist book where they quoted a scientist who was pointing out the simplicity and mindlessness of the way it works. He said, in the quote they used, something about the senselessness of doing the equivalent of leaving a bookcase with a complete set of blueprint and books on building construction in every room in a house. They said that he had just announced to the world that DNA was a blueprint, and a book, and that there was a designer, and that he was a creationist.
A great scientific explanation. For the layman economics, it seems to me it is simple as an analogy with a script for a play. Which is less effort: separating all character lines from the whole and compiling them for each actor individually, or simply photocopying the entire script for everybody and telling them which character they are?
Specifically, in order to cut any genes out, there’d need to be an enzyme for each of them (or maybe a little bit fewer than that), that recognizes it and cuts it out. Each of this huge number of enzymes would need to be coded for by a gene… I’m not surprised that the exceptions either involve the loss of whole chromosomes, which is really easy to do, or involve just genes that would probably be actively harmful outside the germline.
#16: I can think of an alternative scheme: deploy an enzyme that recognizes some subset of epigenetic marks, such as a pattern of histone inactivation of a region of DNA, and slices it out. It would be generic, and would also have the beneficial side effect of removing junk DNA. But since the extra DNA imposes so little cost on the cell, there would be little selective advantage, there might be serious negative consequences (performing surgery on your own DNA is risky), and such a specific enzyme is unlikely to arise by simple chance.
Although I could imagine an enzyme that evolved to extirpate viral insertions having some real advantages, and being coopted to a new function…
I see, that could work…
Somehow, the bdelloid rotifers have gotten rid of their transposable elements. I wonder if they did something like this.
In the early 1980s I used to back up my very important WordStar files (so important that, um, I no longer have them) to a few floppy disks, manually. At some later point (but still so long ago that silicon dating would be required to identify the year), I got an external hard disk big enough that I could drag my entire Applications, Documents and other folders to it, and keep multiple versions of them. This was space-inefficient but boy did it save me time and energy. Nowadays the machine does this automatically, but isn’t there an analogy here?
@Menyambal #14:
I think my initial response to that would have been that some of us are curious and in any case like to read.
Now I want a bookcase in my bathroom. Damn you, Victorian builders, for your stupid oversight!
Trypanosomes actually already do a “DNA surgery” process for their own purposes, despite being single-celled. They have a micronuclei containing scrambled and shuffled genes used for preserving genetic information, and a macronuclei used for transcription of RNA that contains many “chromosomes” each containing one or very few genes.
They use RNA as guides to assemble genes out of the scrambled genes in the micronuclei, rather than epigenetics or specific enzymes. While multicellular organisms don’t do this, it looks like it’s not totally impractical.