So this started with ribose phosphate, glutamine derived ammonia and formate forming a 5-membered ring, then bicarbonate bound a nitrogen and moved to a carbon, aspartate bound the bicarbonate and left its nitrogen behind, another formate bound the same nitrogen as the bicarbonate did before moving and closed a 6-membered ring by binding to the nitrogen. We have a complete purine, IMP.
But IMP is not in DNA or RNA unless made by editing enzymes so this isn’t done. This last stage of purine biosynthesis, stage 5, is 2 parallel sets of 2 reactions that give AMP and GMP. These are equivalent with respect to the origin of life no matter how much I want to make assumptions about which came first. IMP came first if anything, but the surrounding environment may have pushed things towards AMP or GMP (or XMP, below, or things that don’t exist anymore).
I’ll start with AMP synthesis to get the hard part out of the way. The hard part are the P-loops that were mentioned in my last post as one of the oldest groups. I like to write down all of the things and sift out patterns.
Stage 5a.1: aspartate binds again.
Again aspartate is bound by it’s nitrogen, this time to the carbonyl carbon (C=O), and again just a phosphate is needed. More reasons to think about an aspartate accumulation step in molecular evolution. Evolution made things fun and complicated by having a GTP used for this reaction.
The protein that does this is in the A+B3L architecture bin, the P-loop domains-like X bin, and P-loop domains-related homology bin. Finally in the P-loop containing nucleoside triphosphate hydrolases topology bin we find PurA as adenylosuccinate synthetase.
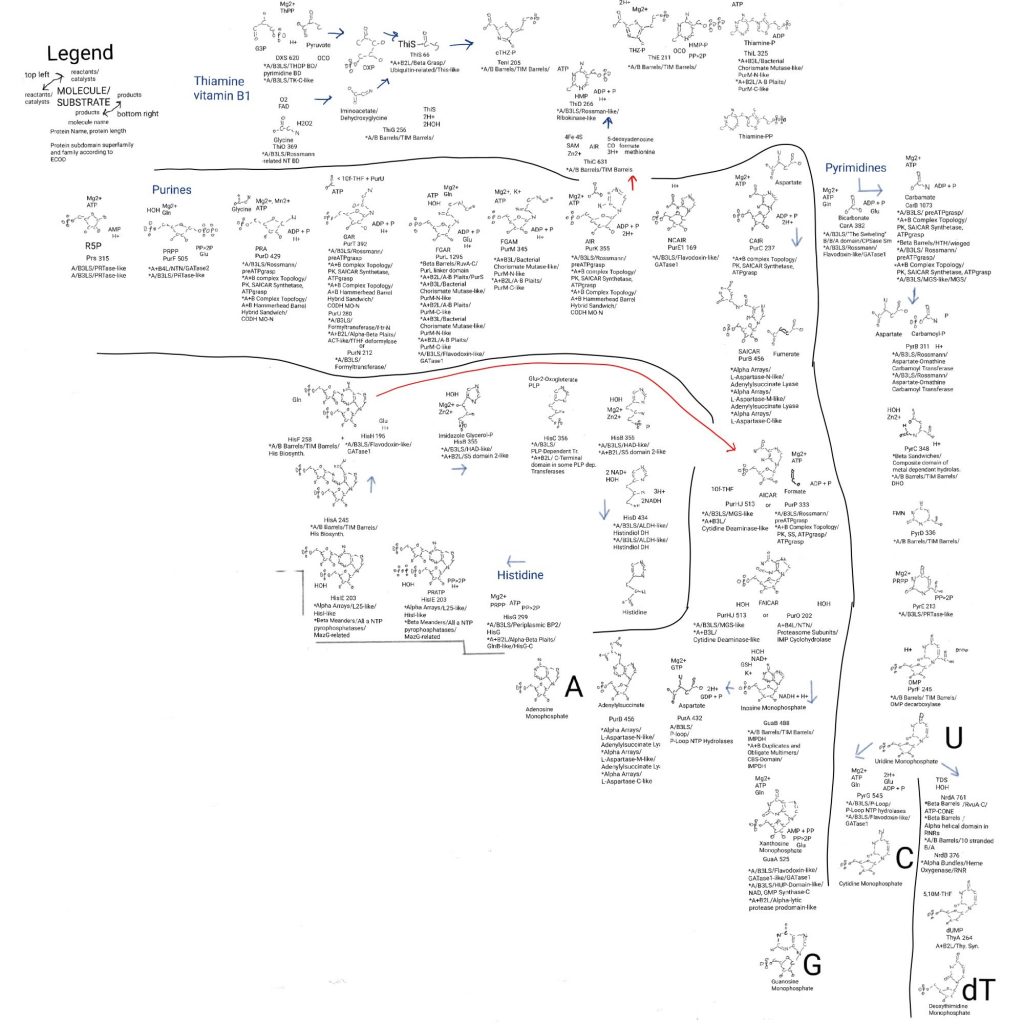
When I click into that topology bin I see why this is the hard one. There are many many P-loop nucleoside triphosphate hydrolases (things that separate parts of nucleotides with water, the phosphates here). It’s worse than the Rossmann domains. And guess what, they might be related.
On the emergence of P-Loop NTPase and Rossmann enzymes from a Beta-Alpha-Beta ancestral fragment. Longo 2020
And that’s what happens when something is an old protein domain, there are lots of kinds in lots of proteins. The following paper is suggestive about the oldest purpose of the P-loops, phosphate control. They looked at P-loops from many proteins and created a minimal fragment that maintains its activity.
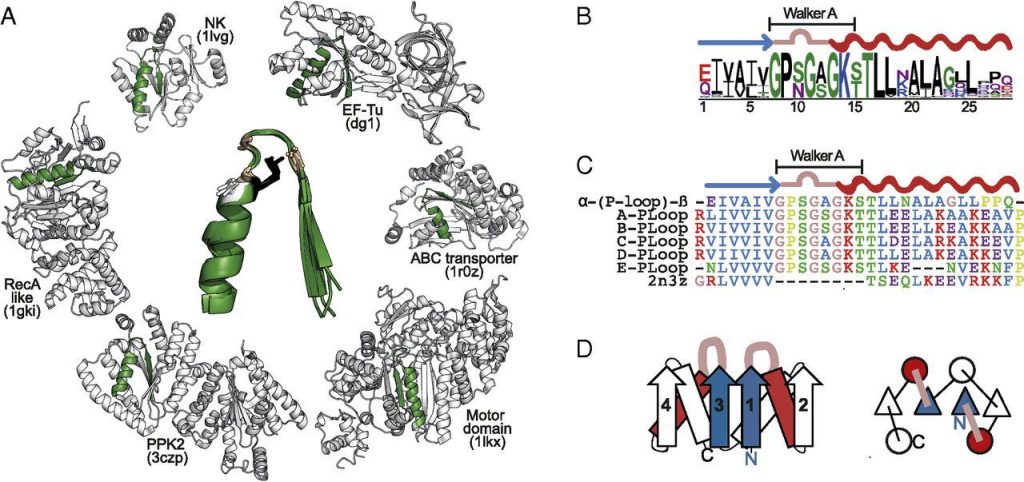
They got it down to 8 amino acid residues.
Simple yet functional phosphate-loop proteins. Romero Romero 2018
Another by the group above looking at Rossmans and P-loops looked at the most ancient domains and found phosphate interaction to be the most basic theme.
There’s little more I can do here, phosphate is so basic and widespread in biology that I’ve little trouble believing every use of phosphate can be in these proteins, active site chemistry, mechanical changes to a protein, changing charge, creating or blocking a binding site…
Still some things occur so often on ECOD that they have repeat family bins with increasing numbers and that is worth mentioning. Sulfotransferases show up repeatedly. So do helicases. Bits of molecular motors dynamin, kinesin, myosin… Iron transport and Iron-sulfur cluster binding proteins.
Polyphosphate kinase 2, Ppk2 seems significant since a store of phosphate is needed for all of this. And there are ATP synthase components.
Formyltetrahydrofolate synthase, FTHFS is what cells use to make RNA since it creates the 10-formyl tetrahydrofolate used to make purines.
It took to almost the end of purine biosynthesis for something truly ancient to show up, and that’s ok because all the rest of purine biosynthesis just needed a single phosphate to work. I can imagine primordial phosphate dispensers accumulating and releasing phosphate in ways where that phosphate can do work without needing a compartment in a protein. A mineral compartment could work fine.
Stage 5a.2: aspartate leaves as fumarate. AMP is synthesized.
It’s PurB again, the same as the last post. That was quick, life was lazy and used the same protein to do it again. It’s worth noting that PurB does not remove aspartate as fumerate in arginine biosynthesis (arginosuccinate).
Stage 5b.1: adding water to IMP.
GuaB has 2 domains. The general reaction is adding water to IMP so it has 2 carbonyl groups, making XMP. Xanthosine monophosphate. What is interesting is that it uses niacin which is based on aspartate and has a biosynthesis pathway similar to pyrimidines. The ring is made and then it is put on ribose to make a proton/electron dispenser/acceptor.
TIM barrels
The first domain is in the A/B Barrels architecture bin, the TIM Alpha Beta barrel X group, and TIM Barrels for homology and topology. This is another hard group because there are so many. This is the 3rd ancient family in these posts after the Rossmans and P-loops. A barrel of 8 repeating A/B segments. Once evolution hit on this it did a lot with it.
TIM barrels confuse me. According to one source I found up to 15 different reactions are catalyzed by this family and it’s the shape that seems conserved.
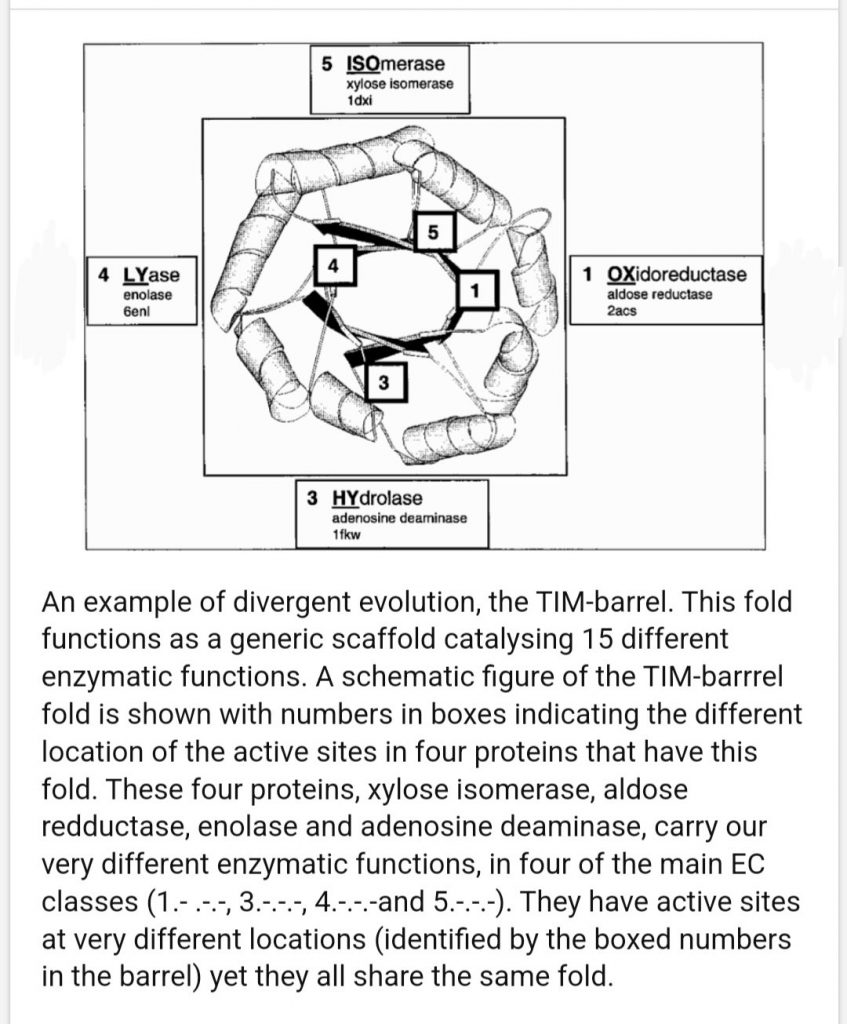
Roles of Specific Peptides in Enzymes. Meroz 2007
Evolution is thought to emphasize the progression from a quarter to a half barrel.
Hidden Sequence Repeats: Additional Evidence for the Origin of TIM-Barrel Family. Ji 2016
So what does shape benefit here? The best I can think of is concentration of things near active sites in that central pore, or multiplication of active sites.
Skip this paragraph if you want to avoid a big list. Significant proteins include: Methylenetetrahydrofolate reductase (makes folate with a 5-methyl), RNAseP, ThiC (makes the big ring of thiamine from purine AIR), Dihydrodipicolinate synthetase (lysine biosynthesis), Quinolinate phosphoribosyl transferase (makes niacin), pyruvate kinase (last step of glycolysis, does a substrate level phosphorylation), glutamate synthase, enolase (glycolysis), Nicotinate phosphoribosyltransferase (adds ribose to the niacin ring), Fructose-bisphosphate aldolase (glycolysis), DAHP synthetase (first step of aromatic amino acid biosynthesis), Type I 3-dehydroquinase (aromatic amino acid biosynthesis), Triosephosphate isomerase (glycolysis, the “TIM” in “TIM barrel”), pterin binding (folate-like), RuBisco large chain (modern photosynthesis carbon acquisition), PdxJ (makes pyridoxal phosphate), ThiG (makes the thiamine 5-membered thiazole ring), Ribulose-phosphate 3 epimerase (makes xylulose phosphate), CO dehydrogenase/acetyl-CoA synthase delta subunit, and lots of things that bind S-adenosyl-methionine.
CBS domain
There is a second domain buried inside of the TIM barrels. This domain is in the “A+B duplicates and obligate multimers” architecture bin, and then the “CBS domain” X, homology and topology bins. According to Interpro CBS domains pair to form single globular domains (called Bateman domains) and GuaB naturally has 2. These domains seem to bind things containing adenylate (adenine) making this a potential regulatory site or a site that binds the adenine of the NAD (niacin) the protein uses to move electrons around.
Stage 5b.2: glutamine provides another ammonia.
GuaA, GMP synthase, works like the other enzymes providing ammonia via glutamine. The ammonia pops off and the phosphate from ATP is used to bind it to the carbon that binds the original carbonyl from IMP. This protein has 3 subdomains.
GATase domain.
This one has been seen before. It’s the GATase1 domain in the Flavodoxin-like bins covered previously. So I’ll refer back to that section.
The next subdomain is new. It’s in the A/B3LS architecture bin, and the “HUP-domain-like” X, homology and topology bins. HUP domains seem to have to do with hydrolysis of the alpha-beta bond of ATP which would leave AMP and diphosphate (the three phosphates of NTPs are labeled alpha, beta, and gamma counting from ribose).
Here’s another list. HUP domains are found in: adenylyl and cytidyl transferases (bind adenosine and cytosine to things, including making niacin). NAD synthase. Glutamine, methionine, lysine, tyrosine, tryptophan, arginine, leucine, isoleucine and valine tRNA synthases. Pantothenate synthase (coenzyme-A), FAD synthase, arginosuccinate synthase, a sacrificial sulfur transferase (LarE, the protein has to be regenerated before it works again), universal stress protein (USP), an Na/Cl/K cotransporter, asparagine synthase, ThiI (thiamine biosynthesis), electron transfer flavoprotein domain, Phosphoadenosine phosphosulfate reductase (PAPS), ATP-sulfurylase and (makes APS which becomes PAPS).
Alpha-lytic protease prodomain-like.
Alpha-lytic protease prodomain is a part of a protein that cuts proteins (protease) that has to be removed before it is active (prodomain). But beyond that GuaA might not be related to anything else in there, it’s in its own homology bin all by itself and is listed as a dimerization domain (so 2 of these proteins interact) making this part done.
And that is it for part 5. Hopefully when I was looking in bins I didn’t ignore anything important. I have 1 more post that puts all of this in a larger context.

Geometry Dash Lite rewards persistence, as players must repeat stages countless times to memorize movement sequences, creating a rewarding feeling when mastering difficult jumps and transitions.
The Greater Gardening of 2026 sounds so futuristic, but it’s all about buying potatoes. I mean, who would have thought that IMP comes first in purine biosynthesis
Geometry Dash SubZero’s chilling graphics, strong beats, and fast-paced courses plunge you into action. Perfect timing and reflexes are essential—one mistake means starting over.