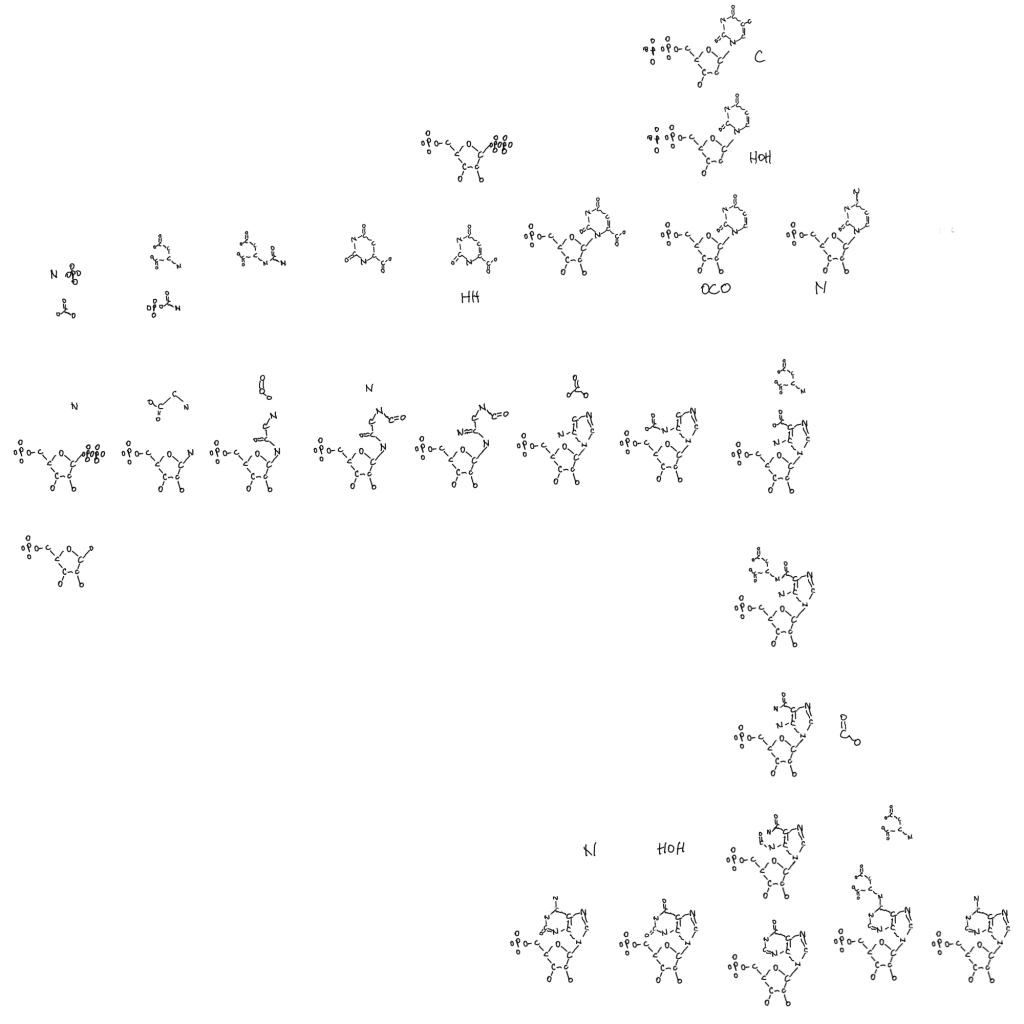
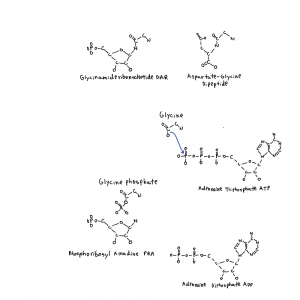
Note: the second figure has been replaced with the correct one, twice. A xylulose bond was in the wrong direction. This is hard.
There are a couple of other origin of life issues that I haven’t focused on yet. One that I was hoping would reveal itself is the issue of “chirality”, or the side of a molecule a bond or atom is on relative to the rest of the molecule in 3D. The strict definition includes things with identical chemical formulas having non-superimposable structures.
I don’t tend to draw in 3D to simplify things. I also don’t include hydrogen to simplify things.
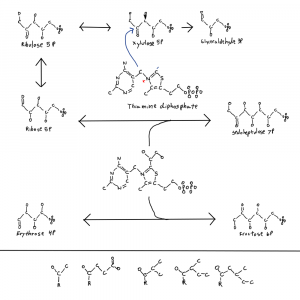
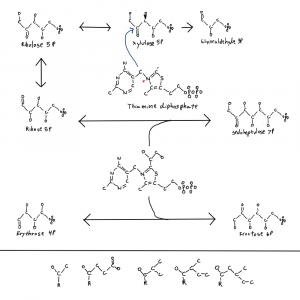
I meant to address this in a previous post where I posted this figure of thiamine and parts of the pentose phosphate pathway. It’s why there is a single hydroxal on xylulose-5P drawn in 3D as if it’s coming at you out of the page. I’ll get to that at the end.
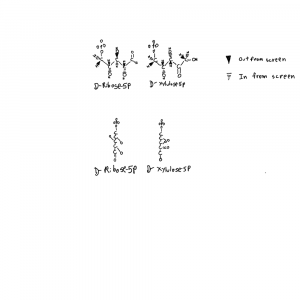
I’ve drawn ribose and xylulose in 3D. Both looking down the length of the molecule and from the side. Carbon actually binds 4 things and is tetrahedral like a pyrimid. So I put the hydrogens back into the upper structures and used conventional dashed and filled wedges to represent bonds going into and out of the plane of the page respectively.
Each of those hydroxals could be where the hydrogen is on the carbon instead. If you flipped the hydrogen and hydroxyl on ribose you can see how the formula would be the same but the structures would not be superimposeable. Things on different sides are designated with R/S or D/L distinctions that refer to single atoms on the structure. D-ribose refers to the stereochemistry around the atom farthest from the aldehyde group in the form of ribose used by life.
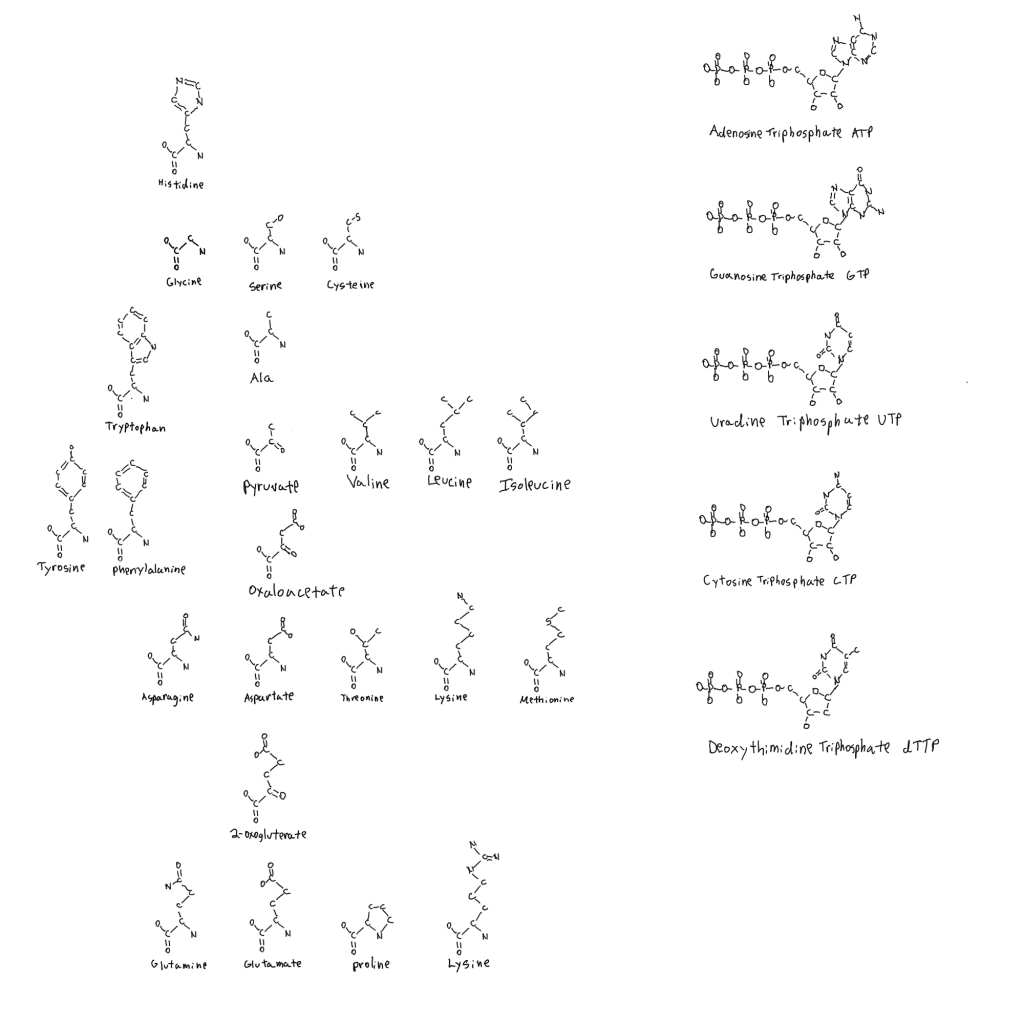
Amino acids are all L- relative to the side chains aside from glycine which has no atoms with stereochemistry.
So why is life one way and not the other when it comes to these bond directions? I don’t know but I noticed that the only sugar with a bond in the other conformation is xylulose-5P so I drew that one bond with 3D on the thiamine figure. The rest are in the other confirmation. And xylulose is the donor for the pentose phosphate pathway 2C molecules. This isn’t a solution to the chirality problem but it is a clue.