Dan Graur has snarled at the authors of a paper defending ENCODE. How could I then resist? I read the offending paper, and I have to say something that will weaken my own reputation as a snarling attack dog myself: it does make a few good points. But it’s mostly using some valid criticisms to defend an indefensible position.
Here’s the abstract.
In its last round of publications in September 2012, the Encyclopedia Of DNA Elements (ENCODE) assigned a biochemical function to most of the human genome, which was taken up by the media as meaning the end of ‘Junk DNA’. This provoked a heated reaction from evolutionary biologists, who among other things claimed that ENCODE adopted a wrong and much too inclusive notion of function, making its dismissal of junk DNA merely rhetorical. We argue that this criticism rests on misunderstandings concerning the nature of the ENCODE project, the relevant notion of function and the claim that most of our genome is junk. We argue that evolutionary accounts of function presuppose functions as ‘causal roles’, and that selection is but a useful proxy for relevant functions, which might well be unsuitable to biomedical research. Taking a closer look at the discovery process in which ENCODE participates, we argue that ENCODE’s strategy of biochemical signatures successfully identified activities of DNA elements with an eye towards causal roles of interest to biomedical research. We argue that ENCODE’s controversial claim of functionality should be interpreted as saying that 80 % of the genome is engaging in relevant biochemical activities and is very likely to have a causal role in phenomena deemed relevant to biomedical research. Finally, we discuss ambiguities in the meaning of junk DNA and in one of the main arguments raised for its prevalence, and we evaluate the impact of ENCODE’s results on the claim that most of our genome is junk.
To simplify it further, they’re arguing that there are different definitions of “function”, and the criticisms of ENCODE rely greatly on the idea that evolution provides the best and most complete test of functionality, while ignoring biomedical functionality.
This is where I shock everyone: I can see that point and even agree with it. After all, if we’re arguing that traits with small effects on fitness are going to be invisible to selection, we have to recognize that there are genetic elements that do have some effect, positive or negative, on the individual, but that aren’t a consequence of selection. So there almost certainly are genetic elements that exist and are relevant to people’s health — they might even be very common — but were never shaped by the effects of selection on the population. An evolution test that identifies elements that have been subject to selection is always going to miss those elements that are invisible to selection.
However, the argument still fails for ENCODE. The authors argue that hypothetically, 80% of the genome has a “causal role in phenomena deemed relevant to biomedical research”. Having an allele that shaves ten years off my expected lifespan may be no big deal from the perspective of evolution, but hey, it matters a heck of a lot to me, so a research program that identified phenomena relevant to my health and well-being is something I would support, even if it didn’t involve evolution.
But did ENCODE do that?
I don’t think so, and the authors don’t point to any specifics. Postulating biomedical causality imposes a whole new set of requirements on the testing, and “a protein sticks to this sequence some times in some cells” is such a loose criterion that it doesn’t help us recognize biomedical relevance, either.
They quote this bit from ENCODE:
The mission of the Encyclopedia of DNA Elements (ENCODE) Project is to enable the scientific and medical communities to interpret the human genome sequence and apply it to understand human biology and improve health.
(ENCODE Consortium 2011, p.1, emphasis added)
Likewise, in the introduction of the 2012 round of papers the authors explicitly frame their enterprise as providing “an important resource for the study of human biology and disease” (ENCODE Consortium 2012, p. 57). This is particularly important because what is relevant to contemporary medicine is not necessarily evolutionarily relevant.
I’ve already said I agree with that in principle. However, it’s just moving the goalposts to yet another position in which ENCODE fails to score. Where, in ENCODE’s assessment of the ‘function’ of sequences, was there an assay to test its effect on human health or biology?
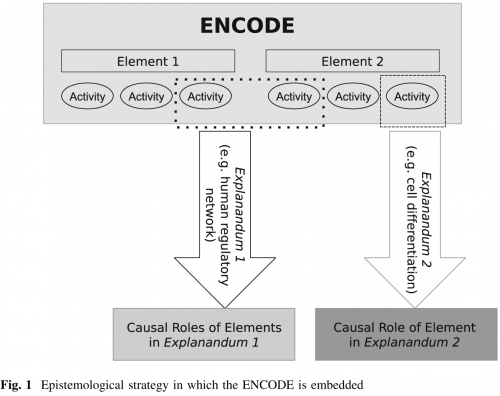
Here’s ENCODE’s definition of function.
Operationally, we define a functional element as a discrete genome segment that encodes a defined product (for example, protein or non-coding RNA) or displays a reproducible biochemical signature (for example, protein binding, or a specific chromatin structure).
Showing that a segment of DNA binds a protein is at least as far from showing a health effect as it is from showing evolutionary significance. The authors are simply floundering about, waving their hands, admitting that ENCODE failed to meet the functional criteria of evolutionary biologists, but maybe it meets the functional criteria of some other discipline, like medicine. Yeah, that’s the ticket, they were helping doctors, or maybe veterinarians, or maybe transhumanists or bioweapons designers. They didn’t actually have any tests for utility in these other domains, but if we fish around enough, maybe we can find an excuse somewhere.
Don’t believe me? Here’s their diagram explaining the phenomenon.
It doesn’t matter what biological activity we’re looking at, there is an ‘explanandum’ under which we can assign it a function. (Philosophers: I’m a bit confused here. Isn’t an ‘explanandum’ the thing that needs to be explained, and an ‘explanans’ the explanation for it, so the terms are a bit off? My pretentious Latin is a bit rusty.)
So even if it’s an unknown explanation, even if the experiment wasn’t designed to test it, even if the generated data doesn’t help solve any of the problems we’re using as a justification, ENCODE’s ass is still covered. How convenient!
Germain P-L, Ratti E, Boem F (2014) Junk or functional DNA? ENCODE and the function controversy. Biol Philos 29:807–831.


Your pretentious Latin is up to the task at hand.
An important issue is how small a selection coefficient has to be before there is no effective natural selection. Theoretical population genetics says that this is roughly when the absolute value of the selection coefficient is less than 1/(4N), where N is the effective population size Human effective population sizes in the paleolithic were small, about 10,000 So the selection coefficient that fails to have much of an effect would be +/- 0.00025. (A selection coefficient is the fractional difference in fitness, so that would be a change of fitness of 2.5 hundredths of a percent). For organisms with larger effective population sizes, such as 1,000,000, the selection coefficient would have to be less than 0.00000025.
Most people’s intuition runs counter to this — we intuitively feel that a 1% difference in fitness shouldn’t matter. But we’re wrong when we think that. If Encode is arguing that having a transcription binding site far from any gene causes a medically significant effect, but not a fitness difference of big enough to matter, well, I’d like to see them make that argument more numerically specific.
Typos: insert period so it reads “the effective population size. Human”
also should read “a fitness difference big enough to matter”
Joe Felsenstein #2
Please, can you provide a ref? I’d be very interested.
(Usually when people ask for refs, they’re questioning the credentials, but here it’s a genuine request, I’d like to use it as a bibliographical starting point).
This effective SelCoefs limit here seems ridiculously small compared to empirical measures (accordingly, these are usually very noisy –most lower SelCoefs are never found to be significant in the statistical sense), I would have been tempted to place the threshold for a (natural) selection effect much above these, and I tend to be a “(closet) panselectionist”. (My bad, but I also strongly feel like a “pluralist”, this is a non-issue to me).
Okidemia
There are many population genetics texts that cover this. My own freely downloadable online text Theoretical Evolutionary Genetics gets to this in Chapter 7, in sections VII.7 and VII.8. Kimura’s 1962 fixation probability formula shows the effect — it’s equation VII-89 and the numerical example in Figure 7.3 uses that formula.
Nielsen and Slatkin’s 2013 book An Introduction to Population Genetics. Theory and Applications covers this in Chapter 8.
You can also try computer simulations. My own lab’s one-locus simulation program PopG is a Java program intended for teaching uses that will work on Windows, Mac OS X, and Linux systems. Alas, not yet on Android or on iOS.
Okidemia,
It’s a common observation that selection coefficients too small to measure in nature, or even in the lab, are still big enough to result in fixation. That’s a problem with our ability to measure, not with selection.
Joe Felsenstein #5
Thanks for pointing to Kimura 1962. I should have guessed actually given that not that many scholars went up because of the academia demographics of the time (not to mention population genetics had to be developped).
I have no doubt when modelling natural selection that fixation is expected even with infinitesimal selection coefficients, I’m actually more wondering as to what are the threshholds or grey zones when drift and selection are at play.
I also find it interesting that fixation is a self tuning off process, given that selection becomes more and more inefficient when advantageous alleles come close to fix in the population. If one imagines an allele suddenly lose its fitness effect, reversion can still go the way back (unless drift comes put an end). The thing that amazes me about evolution, it’s that it is such an open process, it never really closes the door either way. (Well, this is relative of course).
John Harshman #6
Hum. Not entirely convinced. It is a common “prediction”, but not a common “observation” that they reach fix. Given coeff sel distributions, alleles with effects that push fitness closer to “the optimal” will hit fixation first, and I’d tend to think these are still rather the alleles with large to medium effects rather than those at the low distribution tail.
I’m also a bit sceptical that the issue is only our ability to measure and not of selection. If any characteristic (yep sorry, me switching from alleles “theoretical” fitness effect* to measured character based fitness effect may alter the nature of the question) is noisy to measure for us, it is noisy for natural selection as well. The difference is only that NS will have opportunity to make it longer than we do.
Admittedly, I’m also biased in my perception of measuring fitness since my background is on plant ecology, and we’re lucky enough that most measuring is rather easy and we have “good” proxies for fitness for many species (if you don’t precisely choose those that are messier). I should think about how lucky we plant scientists are… :)
*I’m putting sequence based estimates aside by convenience (I have no cue about what these studies tell us ), these might be unequivocal in resolving the debate actually.
Just seen (not read):
http://www.uncommondescent.com/intelligent-design/darwins-defender-pz-myers-remains-unhappy-with-the-encode-findings/
^ Not sure what your point is; Uncommonly Dense is a blog by cdesign proponentsists…
David Marjanović: I think ashley was just bringing it to PZ and the commenters’ attention. (That’s what I interpret based on the “not read” comment: she’s not interested in reading their drivel either, just alerting us to it).
I occasionally find it entertaining to see what rebuttals there are to PZ’s posts out there. They are so often delightfully wrong-headed.
It’s pointing out the creobots/IDiots are happy with ENCODE and their idiosyncratic definition of functionality. As a scientist, I would be ashamed of support from those sources, and would seriously reconsider my null hypotheses and definitions if that was the case. Why doesn’t ENCODE?