How exactly did this dreck, Assembly theory explains and quantifies selection and evolution, get published in Nature?
It’s a stunningly bad paper to be published in such a prestigious journal. Let’s dissect that abstract, shall we?
Scientists have grappled with reconciling biological evolution1,2 with the immutable laws of the Universe defined by physics.
This makes no sense. Evolutionary biologists have not had any problem with physical laws — it has always been assumed, as far as I know, that biology fits within the framework of chemistry and physics. What grappling? Have biologists been proposing theories that violate physics, and they didn’t tell me?
The citations to back up that outré claim are Stuart Kauffman, who can get a little weird but not that weird, and Ryan Gregory, whose papers I’ve used in class, and is probably a bit annoyed at being told his work supports that ridiculous claim.
These laws underpin life’s origin, evolution and the development of human culture and technology, yet they do not predict the emergence of these phenomena.
Sure. Emergent properties exist. We know you can’t simply derive all of biology from Ideal Gas Law. So far, nothing new.
Evolutionary theory explains why some things exist and others do not through the lens of selection.
Uh-oh. Just selection? Tell me you know nothing of evolutionary biology without saying you don’t know anything about evolutionary biology.
To comprehend how diverse, open-ended forms can emerge from physics without an inherent design blueprint, a new approach to understanding and quantifying selection is necessary3,4,5.
Here it comes, more bad theorizing. It is implicit in evolution that there is no “inherent design blueprint,” so where did these authors get the idea that design was a reasonable alternative? They don’t say. This is simply another imaginary controversy they’ve invented to make their theory look more powerful.
We don’t need a new approach
to selection. To support that, they cite Charles Darwin (???) and Sean B. Carroll, and a fellow named Steven Frank, whose work I’m unfamiliar with. A quick search shows that he applies “evolutionary principles to the biochemistry of microbial metabolism,” which doesn’t sound foreign to standard biology, although he does throw the word “design” around a lot.
But here we go:
We present assembly theory (AT) as a framework that does not alter the laws of physics, but redefines the concept of an ‘object’ on which these laws act. AT conceptualizes objects not as point particles, but as entities defined by their possible formation histories. This allows objects to show evidence of selection, within well-defined boundaries of individuals or selected units.
Again, what biological theory has ever been proposed that alters the laws of physics? They keep touting this as a key feature of their model, that it doesn’t break physics, but no credible theory does. This talk of formation histories
is nothing revolutionary, history and contingency are already important concepts in biology. Are they really going to somehow quantify “assembly”? They’re going to try.
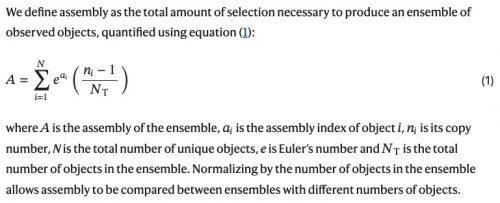
We introduce a measure called assembly (A), capturing the degree of causation required to produce a given ensemble of objects. This approach enables us to incorporate novelty generation and selection into the physics of complex objects. It explains how these objects can be characterized through a forward dynamical process considering their assembly.
I’ve heard this all before, somewhere. A new term invented, a claim of a novel measure of the complexity of a pathway, a shiny new parameter with no clue how to actually measure it? This is just ontogenetic depth! Paul Nelson should be proud that his bad idea has now been enshrined in the pages of Nature, under a new label. I did a quick check: Nelson is not cited in the paper. Sorry, Paul.
Here is all assembly theory
is: You count the number of steps it takes to build an organic something, and presto, you’ve got a number A
that tells you how difficult it was to evolve that something. That’s it. Biology is revolutionized and reconciled with physics. It’s just that stupid.

a–c, AT is generalizable to different classes of objects, illustrated here for three different general types. a, Assembly pathway to construct diethyl phthalate molecule considering molecular bonds as the building blocks. The figure shows the pathway starting with the irreducible constructs to create the molecule with assembly index 8. b, Assembly pathway of a peptide chain by considering building blocks as strings. Left, four amino acids as building blocks. Middle, the actual object and its representation as a string. Right, assembly pathway to construct the string. c, Generalized assembly pathway of an object comprising discrete components.
I told you, it’s just ontogenetic depth, with basic math. Here’s how to calculate A
:
All you have to do is recursively sum the value of A
for each object in the series, and you get the value of A
for the whole! How you calculate the value of A
for, say, acetate or guanine or oxaloacetic acid or your nose or a lobe of your liver is left as an exercise for the reader. It is also left as an exercise for the reader to figure out how A
is going to affect their implementation of evolutionary biology.
By reimagining the concept of matter within assembly spaces, AT provides a powerful interface between physics and biology. It discloses a new aspect of physics emerging at the chemical scale, whereby history and causal contingency influence what exists.
I read the whole thing. I failed to see any new aspect of physics
, or any utility to the theory at all. I don’t see any way to apply this framework
to evolutionary biology, or what I’d do if I could calculate A
for one of my spiders (fortunately, I don’t see any way to figure out the A
of Steatoda triangulosa, so I’m spared the effort of even trying.)
The primary author, Leroy Cronin, a chemistry professor at the University of Glasgow, acknowledges that the work was funded by the John Templeton Foundation. Quelle surprise!
I honestly don’t understand how such a steaming pile managed to get past the editors and reviewers at Nature. It should have been laughed away as pure crank science and tossed out the window. There has to have been a lot of steps where peer review failed…maybe someone should try to calculate the assembly value
for getting a paper published in Nature so we can figure out how it happened.
Sharma, A., Czégel, D., Lachmann, M. et al. Assembly theory explains and quantifies selection and evolution. Nature (2023). https://doi.org/10.1038/s41586-023-06600-9



Quite often in biochemistry, there are multiple pathways to any given compound. So do you just use the simplest one, or average the pathways, or what? And your simplest pathway is going to depend on what starting materials are available.
That was my immediate reaction.
The first sentence of the abstract is wrong.
That is not a good sign.
That’s actually total gibberish. Deepak Chopra level bullshit. Completely meaningless.
This post was absolutely hilarious. X-D
Had me giggling the whole way through.
How to spot flaws in a theory (from “Murder by Death”:
People keep digging up the grave of Intelligent Design.
So they didn’t compute this value for anything?
Now, I don’t feel so bad in never having a paper of mine published in Nature.
They had me at “Universe.”
looks a lot like the partition function from Stat Mech that includes degenerate energy levels of a system.
whywhywhy @7: Someone has done some computin’ based on assembly theory;
Identifying molecules as biosignatures with assembly theory and mass spectrometry
Doesn’t seem completely useless.
That “through the lens of” sounds a lot like humanities’ jargon to me. Nothing wrong with humanities, but a lack of relevant expertise by the authors may explain the poor quality of the article.
The whole “immutable laws of nature defined by physics” thing reminds me of our old friend, “Evolution violates the Law of Entropy”.
Good post! I wonder whether AT is needed to explain how the uniform lsws of physica explain geology. How all the complex fractures of rock in a mountain range arise.
I love the presumption that physics has settled on any answer for anything, and the rest of science needs to conform to it.
No acknowledgement at all that physics is an incredibly dynamic discipline (actually, collection of disciplines) that is constantly revising and refining ideas, discovering new data that are incongruent with currently accepted frameworks, and proposing novel explanations for understanding the physical nature of the universe.
Lot of “immutable” laws are, in fact, quite mutable. As even the most cursory of glances at the last 50 years of physics will tell you.
Man, scientists expressing physics-envy is so weird when the envy is towards some aspect of physics that isn’t present there at all.
Huh. So, the God in the gaps “argument” lives on in a slightly slicker form. I guess now we’ve all been turned into newts, but no one is going to get better
Browsing the abstract, my immediate thought was, “Was this written by ChatGPT?” Sadly, it didn’t seem to ever rise to that level of coherence.
Lot of knee-jerk dismissal here. I thought this Planetary Society interview with Lee Cronin was interesting (starts around 7m 50s);
https://www.planetary.org/planetary-radio/lee-cronin-assembly-theory
@12, you are exactly correct. They are hung up over that old chestnut, the Second Law, which, as an alleged chemist, the principal author should at least understand.
I can’t believe this guy is a chemist: the “interface” between physics and biology is chemistry. Has he never put a few small organic molecules into a flask with a catalyst, shaken the flask once, and seen an enormous macroscale solid, composed of trillions (more, actually) of molecules that instantly self-assembled in an organized, complex, patterned way? Not “designed” at all, but only as a consequence of thermodynamics and kinetics?
Biological systems are just large bunches of atoms, doing their chemical reactions just as usual. I assure everyone within earshot that atoms are just bits of matter than obey the laws of physics, and so are great big bunches of atoms.
IMHO, most chemists are pretty dull, not to say stupid (I am a chemist, so I know), but they usually have a grasp of what is taught in freshman chemistry. I guess that’s unless their bonkers religious views get in the way.
@Rob Grigjanis
It’ll be a bit before I have thoughts on the paper you posted but what computing did they do?
This seems similar to protein folding landscapes resulting from lots of dead ancestor sequences that didn’t make it but applied to all of molecular biology. There’s a difference between “it was likely to turn out this way” and “it was selected” and I don’t see how life is special here.
Maybe it’s semantic distaste of a kind. I don’t see active designers yet. This would be an addition to evolutionary theory, as was pointed out there is more than selection.
Oops, “dead ancestors” is nonsensical. I mean their siblings that didn’t make it.
Do we have to “assemble” the molecules from individual atoms? And those from electrons, protons, and neutrons? Do we count the component quarks, and the (infinite) number of virtual particles that hold all that together?
Do we have to “assemble” the molecules from individual atoms? And those from electrons, protons, and neutrons? Do we count the component quarks, and the (infinite) number of virtual particles that hold all that together?
Sorry for the duplicate post. My assembly function doubled by accident.
No we don’t assemble them, but they do assemble and I chase ghosts in metabolism for a hobby. I think of things assembling due to chemical likelihood.
I’m in assembly of a big figure made up of all of ribose metabolism with a a couple things like pyridoxal phosphate that are cofactors but not ribose related. Each protein and it’s subdomain superfamily information is positioned with it’s substrate molecules.
The 1 and 2 carbon parts are a place I will look for how things may have advanced to more complex chemistry, but it still looks like the Purines are assembled out of lots of smaller molecules relative to other pathways for example.
Design is worse from an implication perspective. It’s got it’s uses but no active designers that I can see. Nested sets of metabolic kludges guided by what was chemically possible and likely.
Here’s how I see the paper so far.
This adds to evolutionary theory at best. I can agree that complex biosignatures can be suggestive of life. And I agree that the pathways I’m looking at were made from parts that could be reused, but were just at useful in the existing systems and not just reused.
Maybe they can find something like a protein folding landscape in molecular space. With CHONPS and HOH. But I’ve been every one of the problems they discussed also discussed in the papers I read. I don’t see any useful metrics yet.
garnetstar @18: Lee Cronin is Regius Chair of Chemistry at the University of Glasgow.
Brony @19: A bit from the paper I linked;
The problem the paper seems to be attempting to address – finding means to “
comprehend how diverse, open-ended forms can emerge from physics without an inherent design blueprint” seems more about the cognitive limitations of the authors and their audience than about problems in biology or physics. Even more so, there seem “intelligent design” red flags in the passage asserting “A hallmark feature of life is how complex objects are generated by evolution, of which many are functional. For example, a DNA molecule holds genetic information reliably and can be copied easily. By contrast, a random string of letters requires much information to describe it, but is not normally seen as very complex or useful. Thus far, science has not been able to find a measure that quantifies the complexity of functionality to distinguish these two cases.” (Dissecting thoroughly how that section is “not even wrong” might well merit a paper in itself.)It seems like the ID crowd finally got a paper (howsoever inept) published in a mostly respectable journal for “Phase I” of the “Wedge Strategy”.
That aside, from a mathematical standpoint the measure seems more useless than defective. In the mathematical theory of computation (which far too ID-iots yammering about “complexity” are familiar with), a formal language corresponds to a string of symbols with a formal grammar of production rules, correspondingly recognizable by computational automata, and with the number of steps in the production roughly corresponding to the number of steps (ergo, amount of time) that the automata must take for recognition. (If you want the fine print on “roughly”, grab copies of Linz and of Sipser and find a computer science professor to explain. Also, while computer science usually works with straight-line strings, that’s not a mathematical necessity; it’s merely less painful for the mathematicians.) So, PZ seems exactly right: “the number of steps it takes”. It looks to me as if the authors may have jumped through a lot of hoops to re-invent time complexity.
I see two big problems lurking here for these ID-iots. First, this re-invention seems to risk leading them eventually to rediscover the Chomsky hierarchy, and realize that the “intelligent design” notion of “irreducible complexity” is BS, in that allowing reducing productions (that drop out an arbitrary chunk) in formal grammars give unrestricted grammars which are necessary enable generating recursively enumerable complexity — loosely speaking, “as complex as can be recognized”. Second, their measure seems defective in that assembling a macro-scale salt (NaCl) crystal apparently would have a high “A” but isn’t what would usually be considered “complex”; the structure is extremely simple. Something akin to Shannon Information and Kolmogorov complexity — counting not the number of steps required to run the program, but how big a program is absolutely required for the automata — seems to give a formal measure of “complex” that aligns better to the concept if given the slightest consideration.
Maybe their measure is only intended to count the steps that use reducing productions; but I don’t think that helps them any.
Sigh; that should have been “which far too FEW ID-iots yammering”.
I don’t see how anyone here see’s utility in this “number.” It has no useful predictive or even organizing value as “number of operations” is poorly defined and does not differentiate for the energetics or probability of the operation. Note that this number would predict I would be more likely to get to the top of a building by leaping than by taking the stairs because the operation count of the first is smaller.
@Rob Grigjanis
I’m there now. Thanks though. I don’t see them invoking designers, just making a lot of mathematical claims with no demonstrated utility.
They can create computational relationships using existing biochemistry that suggest minimal steps and extract a number from it. But what’s the use of their numbers? How do they relate to anything usefully? How do the MA values relate to aspartate here on earth and in an emissions signal from a planet?
We’er already thinking about these things and I don’t see what their numbers do for me.
Brony @32: How could identifying molecules as biosignatures (independently of Earth biology) be useful? Doesn’t that answer itself?
I have been grappling with reconciling biological evolution with the immutable lawmakers of the modern Republican party for half a century. These lawmakers undermine life’s value, economics, and the development of human culture and technology, yet they do not predict the emergence of these phenomena.
I’ve had no success at all. Am I doing something wrong? Do I need even more math and even less attention to boundary conditions?
Can’t help wondering where all the super-skeptics (some of whom are reading ID in their tea leaves, where there is none) were when Angela Collier was butchering high school mathematics in her space elevator video.
@Rob Grigjanis
Because we could already do that and I’m not convinced their number shows that. I don’t see their work providing extra value.
@Jaws
Something like a futile cycle except the way to get something useful out of it is to make it a species-wide concrete example of bad behavior.
https://en.wikipedia.org/wiki/Futile_cycle
At least the original futile cycle makes useful heat.
@Brony:
Where were we already doing that? That’s curiosity, not doubt about your claim. My limited understanding is that we’ve been looking for gases that Earth-like organisms might produce.
@Rob Grigjanis
We have lots of molecules we see biology produce that we haven’t detected out in space. That’s what I mean by we’ve already done it.
I literally don’t see how they know their number mean something is a biosignature when I already see the pathway nature takes to make it. How do you validate this? It’s based on one biosphere and I literally don’t see how their number give me any new understanding, or if they match reality beyond “life make’s this”.
@Brony: Send a letter to NASA.
https://astrobiology.nasa.gov/news/universal-life-detection-astrobiology-assembly-theory/
It really could be that NASA is barking up a horribly wrong tree. I’m nowhere near qualified to say.
@Rob Grigjanis
I’ll pass. NASA can explain how the number is useful. I’m not an expert in assembly theory but it’s up to those experts to show the utility. They have numbers, how do they relate to the rest of the universe? Maybe they can inform a really complicated android game.
Why check a podcast when it’s presumably in the paper?
Because an informal discussion can be informative as to motivation and method, especially when directed at non-specialists?
I’m literally making the choice between the podcast instead of filling in more flavin biosynthesis in my notebook.
It should be in the paper. I’m going to chase my own paredoilia risk now.
https://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-020-01302-7
This paper is like when physicists try to revolutionize biology, except that it’s also ignorant about physics.
The number of steps to assemble a product does not map to the complexity of the product.
The probability of steps does not map to the probability of seeing the final product as there is massive contingency: the abundance of resources and catalysts, the amount of time an assembly can pause waiting for the next step to be possible, the number of alternate pathways and enablers.
The actual map of such assembly may be useful but giving it a number, like 20, conveys no information of any value.
Add to that the contingency tree that can lead to many possible final products. From a given product you can 100% find an assembly path that could get you there, but from the start of the path you can not be so certain of where it will end.
The repeated statements regarding what “physics” is about are generally incomprehensible. Any first-year university physics course will discuss objects having shape and size, objects joining together and splitting apart… It is as though (a caricature of) one corner of the subject, high-energy or “fundamental” particle physics, were taken to be the entirety of the field, and the entire disciplines of statistical and condensed-matter physics were ignored, along with most of the introductory mechanics curriculum.
Citation 3 is to the entirety of Darwin’s Origin, and citation 7 is to the whole of Newton’s Principia. How are nonspecific pointers to old books, in a paper that is not even about the history of science, supposed to communicate anything of value? Citation 22 is to Charlie Bennett’s “Logical depth and physical complexity” and does not support the cited claim. It barely mentions biology at all and makes no statement about some things only arising “late in the evolutionary process”.
“The concept of an object in AT is simple and rigorously defined.”
OK.
“An object is finite, is distinguishable, persists over time and is breakable such that the set of constraints to construct it from elementary building blocks is quantifiable.”
O…K….
“This definition is”
—Wait, that was supposed to be a definition?
“For example, a DNA molecule holds genetic information reliably and can be copied easily. By contrast, a random string of letters requires much information to describe it, but is not normally seen as very complex or useful. Thus far, science has not been able to find a measure that quantifies the complexity of functionality to distinguish these two cases.”
Do they not know that DNA can have a random string of bases and still be copied? Or that DNA can be a homopolymer?
And I’ve created a new evolutionary theory that completely rewrites the laws of physics. It combines protogenetic theory with non-causality to produce combinatory phraseology theory. I’ll be publishing soon, and I expect that CERN will be so impressed that they’ll have to check it out with the Large Hadron Collider to prove my results in biology.
You heard here first. I feel bad that Paul Harvey isn’t still around to pick this up.
Let’s see… Spiders have eight legs, humans have two. Therefore, spiders have a four times larger value for A, and are therefore four times more evolved.
Just gonna leave this here.
Multimodal Techniques for Detecting Alien Life using Assembly Theory and Spectroscopy
‘Multimodal’ just means they use three independent techniques; NMR, tandem mass spectrometry, and IR spectroscopy.
A snippet from the paper linked in #50;
If anyone thinks this claim is wrong, feel free to elucidate.
More crackpottery:
A New Law of Physics Could Support the Idea We’re Living In a Simulation
I checked my calendar and it’s not April. What is going on?
@Rob Grigjanis
How do those extra techniques change the utility of the number? There’s still only the one biosphere using CHONPS and HOH as a data set.
How does using mass spectrometry make change things? I saw that in the paper. What other approaches are they referring to? I see people using mass spec in other places.
I can’t determine if the claim is right. They made a claim about their method based on this biosphere, and I don’t see any effort to show how this number adds anything new to what we’ve already detected in the universe.
People are applying math to protein motifs also and mass spec isn’t really needed to determine if my molecule system is biological.
What does it do?
The kinds of data going into it don’t show me how it’s concretely useful. We have datasets on molecules we detect in space. We have biological pathways that make ones we don’t see in space here on earth.
If we detect something new how does it help? We would look at what we know about how it’s made here and if it could be made in other ways. I see how the number alters what I already know or contributes to deciding if something is life or not.
And how does the number relate to what we have already detected in space? I really want to know.
That should be “I don’t see how the number alters…” in the second to last paragraph.
While these numbers are arbitrarily assigned to useful information (enzyme reaction classification), I’m trying to mentally figure out how the AT numbers will be used practically.
https://enzyme.expasy.org/
Brony @53:
Hm. You could at least read the abstract. They are using independent techniques to test the consistency of their theory.
Srsly mate, if you can’t even be bothered to read the abstract, don’t bother me.
The idea is that other biospheres may not use those as building blocks, but the signature of life would still be a high molecular assembly index, regardless of the elements comprising it. For Earth-based life, the threshhold index (as these folks calculate it) is about 15. Elsewhere, it might be a bit different. What they are doing is finding a way to quantify the complexity of molecules in such a way that it is detectable.
@Rob Grigjanis
Why do I need to stop? You can respond or not.
I did read the abstract. And I honestly don’t see how the separateness of the techniques add independence since there is still no application of the numbers to solving a real world problem.
I just see adding extra complexity to something unnecessarily. I don’t know what 15 means usefully and I’ve still got what we already know to assess future chemistry detection.
PZ was reminded of ontogenetic depth; I was reminded of FSAARs, by Sean Pitman.
Huh; it looks like PZ never posted about fsaars — it was just Sean himself commenting in various threads on ID.
Hey, his website is still up! After 20+ years!
If anyone is interested in reading his attempts to defend his tedious nonsense, I could post the links to the web archives of those threads.
@Reginald Selkirk,
Vopson is off in kookville. At least once before, he’s gotten published in a crap journal and reaped the clickbait. He has claimed that dark matter is information with mass because (waves hands) there’s an E in this equation and an E in this other equation, and what more proof do you need?!
Once I got past the horrible abstract and intro of that paper, I found it to have a bit more meat than you did:
The paper is trying to make a formalized theory that seamlessly joins what I’ll call “physical selection” (because the concept doesn’t actually have a name) with “natural selection” in biology.
Physical selection: The fact that when a new molecule, clump of dirt, or storm forms (any pattern of matter) it will either be stable and stick around (the eye of Jupiter, for example) or it will break apart.
Natural selection: The fact that when a new phenotype emerges, it will either spread through the population or not.
It took FOR EVER for me to understand this from their paper! They suck at writing. That said, their math might be useful for something. I don’t understand how they got published without the paper giving examples of how the theory can be put to practical use.
That said, they did show a practical application in a past paper. Without being told ahead of time, they could determine if a molecule came from a living thing or not, simply based on its Assembly value. They then claimed they could use Assembly Theory to detect life on mars.
@jonperry
I got something like that off of it too which is why I mentioned something like the protein folding landscape for molecules.
https://en.wikipedia.org/wiki/Folding_funnel
I don’t want to say the effort of useless, it’e just there are bits and pieces of that effort in molecular biology already. And it assumes you get what you see with our 4.5 billion year old poly-nucleotide and poly-peptide system. I can see poly-glycine showing up again, but what’s the likelihood of the rest once simple patterns are set?
And I see natural selection as physical already, all those proteins and subdomains splitting off, those molecules are physical and respond to selection.
And it can
Natural and physical. The atomic>molecular level doesn’t warrant special separation to me. And I also don’t really acknowledge a society/biology divide either. It’s biology at a different level of resolution.
@Brony I think the point they’re trying to make is there is no real difference between natural selection and what I called “physical selection” (Dawkins calls it “survival of the stable” in the selfish gene).
They’re trying to say it’s thing from physics to life. I know 2 of the authors are origin of life chemists, which means they might eventually apply this to that research.
The paper claims normal molecules meet Lewontin conditions for evolution by natural selection, but they never said how. I’m baffled that such a horribly written paper was published. My kind assumption is that there was some “curse of knowledge” thing happening in their group. Nobody on the team can explain their ideas to outsiders because they’ve been in a closed loop too long.
They talk about “memory” of a system. They never explain what they mean by that term. The parade of head scratchers in this one is too long to list.
So these
creationists cdesign proponentsests Ontogenetic Depth theoristsAssembly Theory Proponents just want to examine life forms using their metric of “we don’t know so goddidit.” I thought I would cut out as much of the baloney as I could, seeing my triglycerides are too high as it is, but I still can’t stomach it. Hard pass.