I’m getting a bit peeved at all this new technology. Why, back in the day when I was doing electron microscopy work, I’d spend days slicing up tiny fragments of zebrafish embedded in epon-araldite with an ultramicrotome, and I’d end up with hundreds of itty-bitty copper grids that I’d put in the EM one by one, pumping the chamber down to a good vacuum and scanning and focusing and taking pictures. On a film cartridge! That I had to take into a darkroom and process myself! Both ways, uphill, in the snow!
And now look at this. These guys can make a single thin section slice of the whole larva, throw it in a machine, and step back while it automatically scans everything, and then throws it all onto a digital image. Of course, in this case it took their machine 4½ days to shoot over 26,000 images and then stitch them all together, but that’s still far faster than I ever was. I’d slave away to get just one good picture of a chunk of synaptic neuropil maybe 20 micrometers on a side.
Damn. Now I know how John Henry felt.
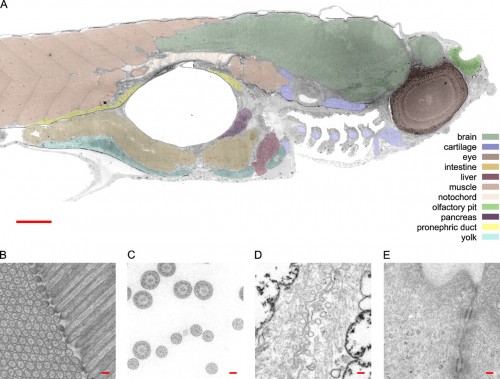
So here’s a transmission electron micrograph of a zebrafish at low resolution, just to help orient yourself. Ah, this is all familiar stuff; I spent most of my time hanging out in the nervous system, but those blocks of muscle (pink) on the left are always beautiful to look at. That big hole in the middle is the swim bladder, and the guts are slung underneath that. This is a parasaggital section — just off the midline — so it slices nicely through the eye (the big dark circle on the right) and also catches one nostril, above (that’s right, not where you might expect it*) and to the right of the eye. The scattered fragmentary stuff at the bottom, in front of the swim bladder, are sections through the pharyngeal arches. Take a look at the pretty cartilaginous rods sliced through in there.
The virtual slide was recorded at 120 kV with a magnification at the detector plane of 9460. A set of points was manually selected to outline the zebrafish and the convex hull of these points was used to define the data collection area. A total of 26,434 unbinned 4k × 4k images was collected with a FEI Eagle CCD camera (>8 s readout time full frame) in 4.5 d. The sample was maintained at −1 µm defocus throughout the whole data collection. The resulting slide of 1,461 × 604 µm2 consists of 921,600 × 380,928 pixels of 1.6 nm square each. The net data content of this slide is 281 Gpixel.
This is at high enough resolution that you can browse around the brain and find synapses and vesicles. Oh, you can’t see that? Go to the visual browser, and you’ll be able to zoom in and in and in. Easily. With no effort. Just glide on in there and find what you want.
Unlike my old experience with EM. Hey, if any of you have a time machine handy, could you grab one of these gadgets and drop it off for me in Eugene, Oregon, about 1982? Thanks.
*In case you’re wondering how nostrils can be above the eye, visualize a bulldog. Now grab it by the snout, and lift upward, stretching the face up so that a forward view is just a shot of the jaws with eyes on either side. Or, better than a bulldog, start with Admiral Ackbar.
Frank G.A. Faas, M. Cristina Avramut, Bernard M. van den Berg, A. Mieke Mommaas, Abraham J. Koster, Raimond B.G. Ravelli (2012) Virtual nanoscopy: Generation of ultra-large high resolution electron microscopy maps. Journal of Cell Biology 198:457-469 DOI: 10.1083/jcb.201201140.

