Next they’ll try telling us the elephant didn’t get its trunk by a crocodile tugging on it.
The genomes of okapi and giraffe have been sequenced, and the signatures of specific genetic changes that are unique to their lineage have been identified. It looks like it wasn’t an act of will after all, but the accumulation of small changes over millions of years. Surprise!
This is an interesting comparison between long-necked mammals, short-necked, related mammals, and mammals as a whole that identified a number of genes that showed evidence of selection. The idea was to find the genes associated with a specific morphological change.
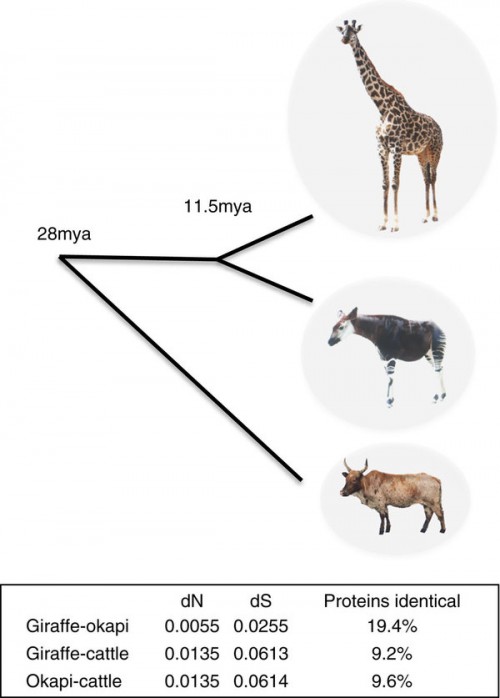
Using the average pairwise synonymous substitution divergence (dS) estimates between giraffe, okapi and cattle as calibrated by the pecoran common ancestor (27.6 mya), the divergence of giraffe and okapi from a common ancestor is estimated to be 11.5 mya.
But of course, there are no such things as genes that uniquely associate with a single trait. A giraffe has a long neck, all right, but that’s also entangled with major changes in the circulatory system, and because they’re adapted to a specific diet, there are all kinds of changes in multiple systems, all interconnected. What they found are 70 genes exhibiting multiple signs of adaptation (MSA), and they’re associated with multiple functions and with each other.
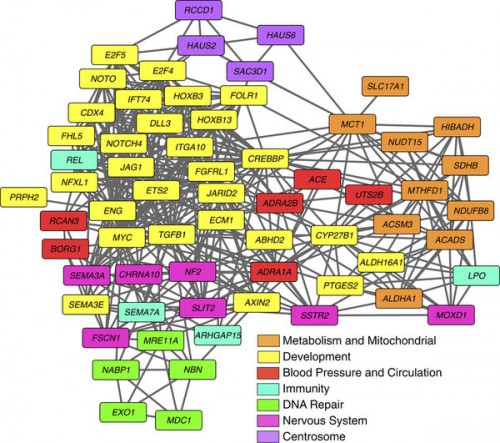
Seventy genes were identified that exhibited MSAs based on amino acid sequence divergence as evaluated by neighbour-joining phylogenetic analysis of mammalian orthologous proteins, enrichment of nonsynonymous substitutions, unique amino acid substitutions at sites otherwise fixed in mammals, substitutions predicted to cause functional changes by Polyphen2 analysis and substitutions under positive selection. Cluster analysis was performed on the set of 70 giraffe MSA genes based on GO Biological Process using Cytoscape 3.0
That’s cool. There are genes that play a role in blood pressure (red), as expected, and diet (orange), and genes of the immune system (cyan; but then, every species sees a lot of changes in its immune system), and in DNA repair (green), which probably has nothing to do with morphology. I am most interested in all those yellow-coded genes, the developmental genes. Unfortunately, “development” is a terrible category which covers a lot of material. It’s rather as if they’d taken all those other, more finely defined categories and just lumped them under “physiology”.
But fortunately, the paper talks a bit more about a few of those developmental genes. One called FOLR1 caught their eye.
The Giraffe FOLR1 shows exceptionally strong evidence for adaptive evolution including six positively selected amino acid substitutions of which two are predicted to cause a significant change in function. FOLR1 mutations are embryonically lethal in mice and produce hypomyelination and neurological defects in humans. In addition to its role in cellular folate transport, FOLR1 is internalized, processed and transported to the nucleus where it regulates components of the FGF and NOTCH pathways. These changes in giraffe FOLR1 may act in concert with similar changes in FGFRL1 and JAG1, components of the FGF and NOTCH pathways, respectively, to forge major developmental adaptations.
FGF, fibroblast growth factor, is a family of signaling genes that play a role in, for instance, cell proliferation — they’re mitogens. They also function in wound healing and angiogenesis, so there is also a connection to the circulatory system. FGFRL1 is also important, because we know that mutations in that gene mess up skeletal and circulatory system development. Notch is a receptor involved in patterning tissues, affecting cell fate decisions and regulating lineages of cells, and has also been implicated in angiogenesis and regulation of bone growth. That’s a lot, so no one can say the problem is close to being solved, but what they’ve found is a few ends to strands of spaghetti that can be be traced deeper into the tangle.
Then there are a few homeobox-containing transcription factors that look promising, and there is an experiment waiting to be done.
We identified three homeobox genes—HOXB3, CDX4 and NOTO—which exhibit significant changes in giraffe compared with other mammals. The advent of gene-editing methods provide a means of testing these hypotheses by introducing the unique amino acid substitutions seen in giraffe into the homologous genes of model organisms and determining the functional consequences.
Yes, please — let’s use CRISPR to put giraffe versions of these homeobox genes into mice. Let’s see if they induce long-necked mice embryos.
Before you get too excited at the idea of mice scurrying along your kitchen floors while browsing the countertops, though, look at the figure of the gene network up above. Putting one or two full-blown giraffe genes into a mouse might produce some measurable morphological changes, but without the integration of all those supporting genes, it’s not likely to be viable. But still, the promise of being able to tease apart gene by gene the changes that produced a giraffe sounds encouraging.
Agaba M, Ishengoma E, and others (2016) Giraffe genome sequence reveals clues to its unique morphology and physiology. Nature Commun. http://dx.doi/org/10.1038/ncomms11519.


I WANT A PET GIRAFFE-MOUSE NOW NOW NOW!
Girouse? Miraffe? Don’t care. I will call him George.
Wouldn’t we expect the most interesting differences to be in regulatory sequences, not protein-coding sequences?
Cue the cdesign proponentsists: They will say that evolution of the giraffe neck is irreducibly complex.
@2: John Harshman
Homeobox genes are regulatory genes specifically associated with development, specifically high level stuff like specifying the length of segments, the number of segments and their identity when they develop further. Transcription factors, which do much of the regulating, are also themselves proteins. The base pair sequences that transcription factors bind to can also change, weakening, strengthening or changing the regulation of the gene the sequence is associated with altogether.
As usual, teh evilutionistas not only has it wrong, backwards, and upside down, but unlogical. The hippopotamus’s neck and legs are not unusually long, its body is simply smaller than made-up-Darwinism requires. Therefore proving, PROVING!, evilutionistiaism is wrong, a line, devil-worship, and Teh Gay!!!!!1!
I’ve petted and fed giraffes at the SF zoo. They have amazing tongues. Maybe these GMO mice will have great big tongues, too.
@4 sharkjack
Yes, I know. By “regulatory sequences” I mean non-coding DNA that interacts with transcription factors, in contrast to regulatory proteins. Those are the elements that seem to change most often to produce developmental changes. They appear and disappear, they become better at binding transcription factors or worse. Most homeobox proteins hardly change at all, which is why mouse Pax-6 works just fine in flies.
So…um….as an Irishman…..who…um….suffers from our well-known curse….um….do you think there might be any….um….ya know…gene therapy sort of thingamajigs….lengthening gene-splicey-mahoney doohickies……ya know…to….emmm….. help a brother out?
I mean, I’m just askin’…..
What about redneck mammals?
Well that graph caption is irreducibly complex. I ran it through Google translate to no avail.
They already did. You can find a summary of Lönnig’s bizarre claims at E&NV.
Wait a minute! This suggests that giraffe necks changed over time, i.e. evolved. That can’t be true. I’ve seen pictures of the Ark in my child Bible story books, and there’s always a hole in the roof for the giraffe to stick its head out. So you guys just stop it with your malarky. God made the giraffe neck long form the beginning. Done!
In case you want to judge yourself Lönnig actually provides download links to two pdf files of his Giraffe book:
http://www.weloennig.de/Giraffe.pdf
http://www.weloennig.de/GiraffaSecondPartEnglish.pdf
Stop it, please, will you? Giraffe doesn’t have a long neck. Its neck is just fine. It’s all the other mammals whose necks are short.
@ 2 John Harshman
I agree with you but its difficult to recognize (and therefore to compare) regulatory sequences strictly on the basis of sequence. Still the paper says they did whole genome sequencing so maybe they’re working on it.
Seconding John Harshman’s questions. IIUC the regulatory switches are typically right next to the coding portion of those genes physically; some (lay?) writeups even include changes in them as changes in that gene. I’m curious if the description here of significant changes refers to large changes in AA sequence.
Where’s the gene you can manipulate for running really fast? Or the gene to make someone really, really smartz? I have heard that you only have to manipulate the genes to make it so….
That is what you think, Mr. Biology Dude. /snark