I’ve now read two novel attempts to explain the existence of junk DNA. To a lot of people, the very idea of junk DNA is offensive: whatever process built us, whether divine fiat or the razor-sharp honing of natural selection, must be powerful, omnipotent or nearly so, and incapable of tolerating any noise or sloppiness, especially not to the degree seen in the eukaryotic genome. There is no room for error in design.
There’s also a strong whiff of human exceptionalism. Look at us, we’re pretty much perfect! Or at least, movie stars and super-models are the pinnacle of creation/evolution. How can you even look at Scarlett Johansson or George Clooney and suggest that they are built of monkey bits and lizard leftovers, or that their manufacture was in any way slapdash?
So there’s an amazing fringe literature out there reaching desperately to find some excuse to justify every scrap of DNA, and especially every bit of human DNA, as purposeful. It sometimes gets weird.
Larry Moran finds an intelligent design creationist who thinks DNA has a fractal dimension. Fractal dimensions are a real thing, and can be calculated; I can’t pretend to understand the math behind them, but given that random noise can have a fractal dimension greater than 1, I can at least say it doesn’t have a necessary relationship to the amount of design information present. Rob Sheldon, on the other hand, claims that he can not only use the parameter as an indicator of how much information is present, but has evidence that humans have the greatest amount of information compression of any species in existence.
The DNA is software. The proteins are the video feed. The nucleus is the CPU. Humans have highly complex coding/decoding machinery in the nucleus. When mathematical analysis is performed on human DNA, it is found to have a fractal information dimension greater than 3 (papers available upon request)–indicating that at least 3 different codes are simultaneously present. This is a number bigger than chimpanzees, whose DNA is not so compressed, and if I recall correctly, come in around 2.5 or so in fractal dimensions. The paper did not analyze onions, but I think it is safe bet that the fractal dimension is < 2.0. What does this changing dimension mean for DNA size? Well the information in DNA is proportional to the volume of phase space, so if humans have dimension 3.0, then the volume ~ (3.2GBytes)^3 ==> 27 GBytes. This dwarfs the 15GBytes of the onion, but then I don’t know the fractal dimensionality of onions.
I know most of you were stopped cold by the bizarre opening metaphor — if you’re like me, you read those first few sentences and were completely paralyzed as you tried to reconcile what you know about biology with that strange analogy, and you were arrested in the process before going on to the greater silliness in the rest of the paragraph. Trust me, it just gets progressively worse, so if you’re still wrestling with the inappropriate comparison of DNA to software, you’re OK, you’re not missing anything.
Here’s the summary, in simple English.
-
I have invented a fudge factor that you multiply the size of a genome by to get it’s true size.
-
I’ve given humans a very big fudge factor, although I’m not going to cite any sources for that. You’ll have to ask me for the papers. (Somehow, I think the author on those papers will be Rob Sheldon.)
-
I vaguely recall that my fudge factor for chimpanzees is somewhat smaller than my fudge factor for humans. Therefore, chimp DNA is less complex than human DNA.
-
I don’t have any data on the fudge factor for onions, but since I have an assumption that onion DNA is simpler than human DNA, it’s a
safe bet
that their fudge factor is smaller than ours. -
Therefore onion DNA is simpler than human DNA, and I have proved it. With math.
Oh, but Sheldon isn’t done yet. He has more nonsense to peddle.
One paper that was published 3 years ago or so, suggested that embryonic development from ovum to embryo was driven by a clock. As the transcriptase zipped along the DNA, proteins were made successively by the cell, and the ordering and timing of the proteins were such as to drive the embryogenesis and development. In other words, the spatial location of the DNA was converted into temporal development of the organism. Then if an organism needed to prolong a stage of embryogenesis, the most direct way would involve adding more DNA. No extra machinery is needed, no added complications and regulators, just another 1GByte of DNA to transcribe and the necessary 30 minutes will be added to the development.
Congratulations, Rob Sheldon! You have reinvented Haeckel’s Law of Terminal Addition!
Alas for Sheldon’s Law of Terminal Addition, we already know it’s false. There is no simple sequential arrangement of information in the genome that corresponds to timing or spatial organization; the chromosome is not a map. We also know that the order of genes is greatly scrambled between different species — the human genome is not the mouse genome with a bunch of extra stuff tacked on.
There are exceptions, and I think Sheldon is regurgitating a badly muddled explanation of Hox gene clusters. Hox genes in many (but not all!) animal species are organized linearly in a cluster, ordered both from front to back and from early to late (here’s an explanation by some bozo in Nature Education).
This is, however, not a universal explanation for how all spatially localized proteins are ordered in the genome. It is also not correct that it’s handled by a transcriptase clock, an idea that makes no sense whatsoever. The regulation of Hox genes is really, really complicated, not just a matter of taking longer with longer sequences.
Do I also need to point out that if it’s all just a matter of timing, and that more complex species will require longer times to develop, then giraffes, elephants, zebras, and whales must be more complex than humans because they have significantly longer gestation times, and we’re just slightly more sophisticated than moose?
But before you take away the lesson that it’s just creationists who are idiots, there are also quite a few serious scientists who have equally silly ideas, and get published in serious journals. T. Ryan Gregory finds a paper that explains the function of junk DNA, published in Molecular Plant by someone named Freeling. Gregory cruelly calls it “word salad about junk DNA.” But that can’t be! It’s published! In a real journal! You’ll have to judge for yourself.
The Genome Balance Hypothesis originated from a recent study that provided a mechanism for the phenomenon of genome dominance in ancient polyploids: unique 24nt RNA coverage near genes is greater in genes on the recessive subgenome irrespective of differences in gene expression. 24nt RNAs target transposons. Transposon position effects are now hypothesized to balance the expression of networked genes and provide spring-like tension between pericentromeric heterochromatin and microtubules. The balance (coordination) of gene expression and centromere movement are under selection. Our hypothesis states that this balance can be maintained by many or few transposons about equally well. We explain known, balanced distributions of junk DNA within genomes, and between subgenomes in allopolyploids (and our hypothesis passes “the onion test” for any so-called solution to the C-value paradox). Importantly, when the allotetraploid maize chromosomes delete redundant genes, their nearby transposons are also lost; this result is explained if transposons near genes function. The Genome Balance Hypothesis is hypothetical because the position effect mechanisms implicated are not proved to apply to all junk DNA, and the continuous nature of the centromeric and gene position effects have not yet been studied as a single phenomenon.
I can usually pride myself on my ability to explain complex papers reasonably simply (or simpler, anyway), but I find myself completely stumped by that paragraph. So junk DNA, which is synonymous with transposons, acts as springs between regions of DNA and…microtubules? What? And a balanced distribution of these springs is required by selection? Why? I don’t know. I give up.
Let’s turn to the figures in the paper and see if they explain anything.
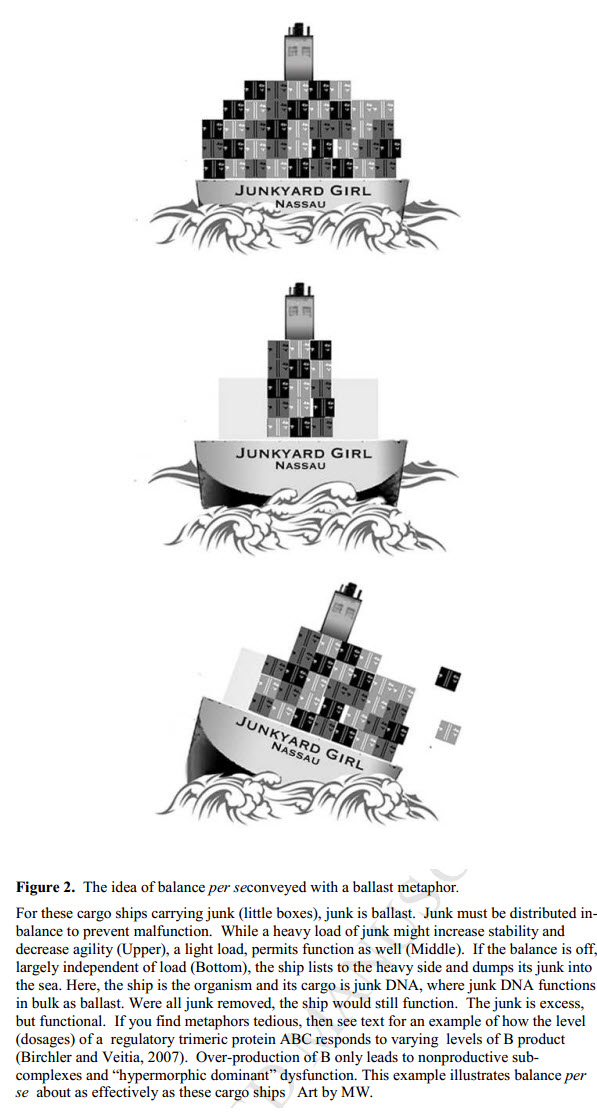
Welp, nope. Now the springs have turned into ballast on a boat, but we have no explanations for why the genome needs any amount of “ballast”. Or springs. Or what it means to have a genome capsize. Or where the boat is taking that load of ballast.
I have an idea. What if we accept the idea that humans are animals, no more exceptional in their evolution than mice, or giraffes, or moose. What if we also accept that chance plays a significant role in evolution, as does selection, and that our genomes are a contingent product of a combination of processes with no overarching design — that the operational outcome of the whole shebang is generational expediency, that all Nature expects is that we’re good enough to survive, and that if we’re a little bit better at surviving than others, our lineage will thrive at their expense.
It’s a very simple concept.
And, the thing is, it explains a lot of phenomena that are difficult to justify with a flawless and efficient designer, and it fits all our observations of genomes and organisms remarkably well.
The only problem is psychological: you do have to kick humans off that throne you’ve put them on, and you also have to evict that invisible magical benevolent designer out of the house. It’s no loss, though, since the invisible designer never seemed to do anything, and humans work better when they aren’t lounging about on thrones.


Yes, “fractal dimension” can be calculated for mathematical fractal objects—and had been, in effect, long before Mandelbrot coined that particular name for the concept. But there is no physical object that is at all honestly modeled by a mathematical fractal! (Physical “random noise” is not a counterexample: your favorite mathematical model of physical “random noise”, say Gaussian noise, is a convenient idealization of physical noise, and it works well, but they’re not identical.) The mathematical definitions of fractal (Mandelbrot’s own, and others since) are all variations on “self-similarity at infinitely many different scales that tend to 0”: the purported physical examples of fractals (like, most famously, the coastline of England) are rarely self-similar at more than a small handful of different scales. How could it be otherwise, when energy is quantized and space might as well be (and maybe is) in the sense that there is a minimum resolution in observation of spatial separation?
If order of reading was important for genetic information, I would not be here. I’ve got a gene translocation, but am nonetheless (arguably) phenotype normal.
I know a bit about fractals, and fractal dimensions are applicable to geometric constructs. I see no way to correlate them with an information density. To even begin to speculate about such a relationship, one would need to know a priori the level of redundancy in the DNA and the level of noise. One cannot prove anything from his approach. Sheesh.
And as leerudolph has noted, DNA is not self-similar as one changes scale. The whole approach looks totally bogus.
I am pretty sure that is not how math works.
“The DNA is software. The proteins are the video feed. The nucleus is the CPU. Humans have highly complex coding/decoding machinery in the nucleus.”
You know this doesn’t actually work as an analogy with a computer, right?
The DNA could be the software, sure.
Then the RNA polymerase is the closest thing you’ve got to a CPU (i.e. the thing that interprets the machine instructions)
The proteins become the actions of the CPU (read from memory, write to device, add, subtract bytes etc.) I don’t think this works at all, but you guys are the biologists, not me.
A “video feed” has no analogy in biology: if you had a “video feed”, it would mean taking basically raw data from your DNA, encapsulating it in something analogous to an Ethernet packet, then sending that packet to another device that could strip off the Ethernet and reconstitute the DNA. Maybe the standard copying of DNA is like that, but I’m willing to bet (based on some vague stuff I’ve read about DNA replication) that the analogy will fail pretty hard.
The only thing that comes close to that is some stuff I read once (possibly B.S.? I don’t know) about how trees can communicate with each other, through airborne somethings, when they figure out how to defeat pests. *IF* such a mechanism exists, it would be the closest analogy to a “video feed”.
a) This guy seems to have lost his calculator
b) He put the GB (GiB?) inside the parentheses, so the result is 32.7 cubic gigabyte.
I have no idea what a cubic byte is, but i’m sure you can’t just compare it to bytes.
It’s like saying 2m^3 > 1m. Nope. It just doesn’t work that way.
And ballast is not carried above the water line. Useless weight above the water line is just a hazard.
Since the analogy is full of crap, I can feel contentedly smug in my lack of knowledge about all that fractal stuff.
As a code monkey with bioinformatics experience, I fucking ~hate it~ when people use computer programs as an analogy for DNA. I could tolerate a comparison to something like a probabilistic interrupt-driven architecture, but by that point you’re so far away from the dominant computing paradigm that it’s not a useful explanatory device anymore.
Also hate when computer-brained geniuses try to grok biological evolution via genetic algorithms. Talk about the tail wagging the dog.
In case anyone is curious about it, fractal dimension isn’t too exotic to understand on an intuitive level. If you take a line segment and double the scale, there’s 2 times as much line there. If you take a square and double the scale, there’s 4 = 2^2 times as much, and if you take a cube there will be 8 = 2^3 times as much.
If you take a Sierpinski triangle and double the scale, you can see that now you have three copies of the thing you started with, so there’s 3 times as much there. So it’s somewhere between the line and the square; 3 = 2^1.58… so it has a fractal dimension of about 1.58. All the rest of the math is just extending the concept to cases where you don’t get exact copies.
It’s a neat idea with all sorts of uses. This is of course not one of them, because it’s based on the confused assumption that phase space tells you coding information. Information in a mathematical sense doesn’t have to be useful, it just has to be hard to specify; a random sequence of symbols actually has much higher information content than a regular pattern.
It doesn’t make any sense why that would be the issue to consider. From what I can tell, most genes look like random sequences of nucleotides; the only logic behind them is what they happen to cause. At the same time, finding a sequence is random is not a good argument it isn’t junk. And yet I’m not sure you can argue the opposite either; does anyone expect junk DNA to be purely random? It’s non-functional so should be randomized over time, but if it turned out there was some partly-repeated pattern from broken fragments of genes, switches, and viral insertions, I’m not sure who would be surprised.
Yes, I know we know this is all garbage…I just thought I might join leerudolph in criticizing more than just the analogy, because there’s lots more here that deserves it.
a cubic byte ::= voxel… { kinda. voxel is literally the 3d version of a 2d pixel. but ‘close enuf’}
to highlight:
And that is what SCIENCE is all about, i.e. : Coming up with a bizarre explanation for a poorly understood phenomena, then publishing it, to invite Discussion. (so long as one is able to relinquish said notion when presented with counter-evidence or a better explanation…)
The paper that Rob Sheldon seems to be referring to is:
Garte, Fractal properties of the human genome, Journal of Theoretical Biology 2004
The only information the paper deals with is the pattern of coding vs noncoding. The abstract is in direct contradiction with Rob Sheldon’s account.
Abstract:
That’s not a bozo, that’s a poopyhead!
I gotta agree with taco_emoji here. I code and I do biology. But I do more biology than code. So when coders talk like they know something about biology it shits me to tears. Biology might be considered a form of computation, as can the universe, but these designed silicon boxes are not doing the same sort of thing. Analogies can be drawn but the map is not the territory. And this is the fundamental problem with analogies; they result in conflation and often at the expense of understanding, rather than being instructive. Most people who understand biology through a silicon computation metaphor are hopelessly lost when it comes to understanding how biology really works. Methinks our friend Rob Sheldon is an example of this.
@nomadiq
That’s the alarming thing to me. We have a person who apparently doesn’t understand biology (according to biologists I trust, I can’t be sure myself) trying to draw an analogy with computers (which *I* can tell he clearly doesn’t understand).
I mean, how do you make an analogy when you understand neither side of the metaphor?
Yeah.. If DNA was a “program”, then it would be sort of like HTML – it bloody works, sort of, no matter how much useless crap is in it, or how broken the code. Something will still come out the other end, regardless of how badly some idiot screws up the “code”. This is in contrast with “program languages”, which are intended not to be information on how to make something, i.e., display a page, or build a human, but the “thing itself”, where fucking on one semi-colon can cause it to either totally fail to work, at all, or implode on itself. Mind… there may be elements of “both” sort of “code” in genetics, but most of it seems to be more like a bloody web page, and yeah, even when there is the “language as the thing” sort of code in a page, usually it only breaks the bit that relies on that bit, while the rest works – the equivalent of breaking the DNA that, say, produces a skin pigmentation, but nothing else of any sort.
Pretty sure.. fractals like.. don’t work if you break the math some place mid-computation, or what ever, too…
But, yeah, what #14 says.. before you can try to make analogies, you have to know what the F you are talking about, with respect to “both” subjects, but then… that would preclude making stupid analogies that rely on things working like you imagine they do, instead of the reality that they bloody don’t. They might, almost, have a sane analogy if they had been talking about some strange esoteric computer language, with rules that where intentionally made up to see if it was even possible to write something in it, and have it work, but with no intention for it to make any damn sense, at all, from the standpoint of either design, reliable function, or usability. But… nah, that would have pretty much ruined their analogy… lol
The concept of fractal dimension can be applied to DNA sequences. However, the results (a) haven’t been astoundingly revolutionary, (b) support the idea of junk DNA and (c) go in the opposite direction to what Sheldon claims.
The basic idea is that you can transform a DNA sequence (“ATGAAACGCAT…”) into a geometrical object. Associate each of the four bases with a direction, e.g., A = up, T = down, G = left, C = right. Then, a DNA sequence becomes a series of instructions for taking a walk on a 2D grid. The shape of this walk can then be analyzed statistically.
People tried this on the yeast genome (if you’ve got the sequences and you have a computer, why not?, I suppose). The upshot was that the fractal dimension of coding DNA was smaller than that of noncoding DNA.
I’ve seen a few variations on this general idea, none of them mind-blowing.
We have a replacement for the clause “because reasons”. I’m now going to use “because fractals” for all Woo statements now.
Fractals, the new quantum.
My expertise is in software, not biology. I can’t even think of anything to say about the “video feed” analogy. And I don’t think it quite makes sense to store the software inside the CPU. Even if that “transcriptase clock” idea was correct, it seems to me that the junk DNA would then be like using Sleep() statements to fix timing issues in multi-threaded apps (i.e. we don’t know why this code doesn’t work, but if you add a “pause for 1/10 of a second” in this spot, the problem goes away.)
If you’re going to use the computer analogy seems like junk DNA might be more like cookies that are floating around from unused or long dead web sites and if MySpace ever makes a comeback some of those cookies will be ready to express themselves.
I know it’s a stretch but I am statistcian not a biologist or programmer
Initially, I read “video feed” as “video tape”; that all the “bits” of data are stored _on_the_tape_ as little magnetic dots (N=1, S=0). Then, reading the whole “analogy” holistically, the whole metaphor crashed into nonsense.
Fuck all. Next question, please.
To reiterate a previous comment:
Moran does NOT know how to do even basic mathematics, “fractals” is way beyond his skillset.
I.E. : He wrote, “(3.2GBytes)^3 ==> 27 GBytes” Even allowing his misplacement of the parens, and mistaken inclusion of the order-of-magnitude as mere typos: (3.2)^3 == 32.7; where 27 isn’t even close. Moran, Giga- = 10^9, (10^9)^3 = 10^27; so (3.2*10^9 bytes)^3 == 32.768*10^27 cubic-bytes; as in (2 inches)^3 == 8 cubic inches.
Moran, if you’re going to try to dazzle us with advanced mathematics, at least get your simple arithmetic correct!!! Else, you’ll never be taken seriously and get your name denigrated to Moron.
When I saw “I’ve now read two novel attempts to explain”, I somehow expected this to be a book review.
The quote is from .
Hey twas brillig, if you’re gonna be calling people morons maybe you should first learn to read, hm?
David, and Island, got me. Yes I was too quick with that insulting name-calling. My reading is sometimes too quick for full integration. I saw Moran mentioned at the beginning of the paragraph introducing the blockquote, and missed the intermediary sentence attributing the quote to Sheldon.
mea culpa Apologies to all for ‘~issing into the wind’ (instead of downwind). I seem to have got that egg all over my own face. Just read the maths part of that note of mine and disregard the insults at the end.
Yeah, my teeth would beg to differ.