I’ve had, off and on, a minor obsession with a particular number. That number is 210. Look for it in any review of evolutionary complexity; some number in the 200+ range will get trotted out as the estimated number of cell types in a chordate/vertebrate/mammal/human, and it will typically be touted as the peak number of cell types in any organism. We have the most cellular diversity! Yay for us! We are sooo complicated!
It’s an aspect of the Deflated Ego problem, in which scientists exercise a little confirmation bias to find some metric that puts humans at the top of the complexity heap. Larry Moran is talking about the various techniques people use to inflate the complexity of the genome, making special case arguments for novel molecular gimmicks that we mammals use to get far more ooomph out of our genes than those other, lesser organisms do.
As I was reading it, I had this sense of deja vu, and using my psychic powers, I predicted that someone was going to make the argument that because we mammals have so many more cell types than other organisms, there must be some genetic trick we’re playing to increase the number of outcomes from our developmental processes, and that therefore there must be something to it. Because we are measurably more complex than other animals, there must be a mechanism to get more complexity out of our 20,000 genes than nematodes get out of theirs.
And did I call it? I did. Very first comment:
I dont think its a sign of an inflated ego to think mammals are more complex than flies. There are objective measures one could use such as cell type number, number of neurons or neural connectivity.
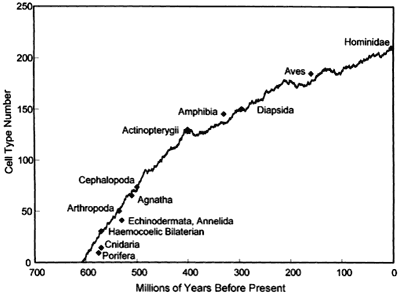
There’s a problem with this claim, though. Many people, including quite a few prestigious scientists, believe that cell type number in various organisms has actually been measured, and you’ll even find respected people like Valentine putting together charts like this:
That chart is total bullshit. You know how I expressed my visceral repugnance for an MRA who made up a “sexual market value” chart? I feel the same rage when I see this chart. There is no data supporting it. There we see humans listed as having 210 cell types, and everything else is lesser: birds have only 187 cell types. Do you believe that? I sure as hell don’t.
I periodically get a bit pissed off about this. I wrote about it in a thread on Talk.Origins in 2000, and I’ve put a copy of that below. I complained about it in a blog post from 2007. It hasn’t sunk in. I still run into this nonsense fairly regularly.
The short answer: this number and imaginary trend in cell type complexity are derived entirely from an otherwise obscure and rarely cited 60 year old review paper that contained no original data on the problem; the values are all guesswork, estimates from the number of cell types listed in histology textbooks. That’s it.
The long answer, my digging from 13 years ago:
This is a topic in which I’ve long had an interest, of a peculiar and morbid sort. It’s been a case of occasionally running into these arguments about cell types, and wondering whether I’m stupidly missing something obvious, or whether the authors of these claims are the cockeyed ones. I can’t see a middle ground, it’s one or the other. Maybe somebody here can point out how idiotic I must be.
The issue is whether we can identify a good measure of organismal complexity. One way, you might think, would be to look at the number of different cell types present. I first ran across this metric in the late ’70s, in JT Bonner’s book _On Development: the biology of form_. He has a number of provocative graphs in that book, that try to relate various parameters of form to life history and evolution. Some of the parameters are easy to assess: maximum length, or approximate number of cells (which is just roughly proportional to volume). Others were messy: number of different cell types. Bonner didn’t push that one too much, just pointing out that a plot of number of types vs. total number of cells was sorta linear on a logarithmic plot, and he kept the comparison crude, looking at a whale vs. a sequoia vs. a sponge, that sort of thing. He also said of counting cell types that it was “in itself an approximate and arbitrary task”, but doesn’t say or cite where the numbers he used came from, or how they were obtained.
It came up again in Stuart Kauffman’s work. He tried to justify his claim that the number of cell states (or types) in an organism was a function of the number of genes, and he put together a chart of genome size vs. number of cell types. It was glaringly bogus. He (or someone) clearly selected the data, leaving out organisms with what I guess he would consider anomalous genome sizes — and Raff and Kaufman thoroughly trashed that entire line of argument in their chapter on the C-value paradox in _Embryos, Genes, and Evolution_, showing that one axis of Kauffman’s graph has to be invalid. Nobody has touched on that other axis, the number of cell types, and I’m still wondering how anybody determined that humans have precisely 210 different kinds of cells, while flies have 50 (those numbers seem to have become canonized, by the way — I’ve found several sources that cite them, +/- a bit, but very few say where they came from).
And then Morton mentions this interesting little paper that I hadn’t seen before:
Valentine, JW, AG Collins, CP Meyer (1994) Morphological complexity increase in metazoans. Paleobiology 20(2):131-142.
[note to Glenn: the citation on your page is incorrect. It’s in Paleobiology, not Paleontology]
Abstract.-The number of cell types required fo rthe constructon of a metazoan body plan can serve as an index of morphological (or anatomical) complexity; living metazoans range from four (placozoans) to over 200 (hominids) somatic cell types. A plot of the times of origin of body plans against their cell type numbers suggests that the upper bound of complexity has increased more or less steadily from the earliest metazoans until today, at an average rate of about one cell typer per 3 my (when nerve cells are lumped). Computer models in which increase or decrease in cell type number was random were used to investigate the behavior of the upper bound of cell type number in evolving clades. The models are Markovian; variance in cell type number increases linearly through time. Scaled to the fossil record of the upper bound of cell type numbers, the models suggest that early rates of increase in maximum complexity were relatively high. the models and the data are mutually consistent and suggest that the Metazoa originated near 600 Ma, the the metazoan “explosion” near the Precambrian/Cambrian transition was not associated with any important increase in complexity of body plans, and that important decreases in the upper bound of complexity are unlikely to have occurred.
At least, the paper *sounds* interesting. After reading it, though, I’m left feeling that it is an awful, lousy bit of work.
The first major flaw: there is no data in the paper. The first figure is a plot of cell type number against age, in millions of years before the present — the numbers and groups described are listed on Glenn Morton’s page. These are the observations against which several computer models will be compared. These data were not measured by the authors, but were gleaned from the literature. The sources for these critical numbers are listed in an appendix, about which more in a little bit.
The bulk of the paper is about the computer models they developed. The final figure is the same as the first, showing the data points from the literature with the plot generated by their best-fit simulation superimposed. It’s a very good fit. From this, they make several conclusions: 1) that their model is in good agreement with the historical data, 2) that the rate of increase in complexity was greatest near the origin of metazoans, 3) that that origin was relatively late, and 4) there was no particular change in rate during the Cambrian explosion. It is a fine example of GIGO.
The work is completely reliant on the validity of the data about cell type number, which is not generated by the authors, and worse, which is not even critically evaluated by the authors. It is just accepted. That data left me cold, though, with lots of questions.
What is a cell type? There was no attempt to define it. Histologically, it’s a fuzzy mess — you can go through any histology text and find long lists of cells types that have been recognized by morphology, location, staining properties, and so forth. I just skimmed through the index of an old text I have on hand (Leeson and Leeson), and without trying too hard, counted a bit more than a hundred distinct, named, vertebrate cell types in the first 5 pages…and there were 25 more pages to go. What criteria are the authors using? How well do these superficial criteria for identification mesh with the molecular reality of the processes that shape these cells?
Why did they throw out huge categories of cells? The nervous system is simply not considered — it’s ‘lumped’. This seems to me to be grossly inappropriate. Here is this HUGE heap of cellular diversity, in which half the genome is involved, and it is discarded in what are supposedly quantitative models. I can guess that it was thrown out because it is impossible to quantify…but that doesn’t sound like a good excuse if you are trying to model numbers. Furthermore, they only count cells in adults, so cell types found only in larvae or juveniles are rejected. Whoops. Isn’t that an admission that complexity in arthropods is going to be seriously underestimated? I don’t know, since they don’t say how they define a cell type.
How did they get these tidy single numbers for a whole group? ‘Arthropods’ have only 50 cell types. They admit that “within some groups there is a significant range of cell type numbers”. The range of variation, however, is not reflected in any of their graphs, nor which groups exhibit this range. Instead, they say, they picked a representative “primitive number” of cell types from “the more primitive living forms within each group”. I guess the more primitive living forms haven’t done any evolving.
A really bothersome and related point: the high end of their plot is anchored by the hominids, with 210 cell types and a time of origin within the last few million years. Remember, they are going to fit all these computer-generated curves to these data, and they explicitly scale everything to this endpoint and an earlier one. This point is invalid, though. We humans don’t have any novel cell types that were generated a few million years ago — that number of 210 cells ought to be applied to all of Mammalia, and the time of origin shoved back a hundred million years. Or more. Is there any reason to think 200 million year old therapsids were lacking any significant number of histological cell types found in mammals today?
For that matter, why should we think that these cell type numbers are anything but arbitrary indicators of the relative amount of time histologists have spent picking over the tissues of these various organisms? Do fish really have fewer cell types than mammals, or just different ones? Fish may lack all the cell types associated with hairs, but we don’t have all the ones that form scales. The authors show amphibians as being more complex than fish, on the basis of cell type counts in living forms…and that is completely the reverse of what I would expect, if I thought there was any difference at all.
What was really the killer for me, and what I was really looking for, was the primary sources for these numbers. These are listed at the very end, in a separate appendix. A few are easy: it’s not hard to imagine being able to count all the different cell types in a sponge or a jellyfish. One is admitted speculation by Valentine — he estimates the number of cells a primitive hemocoelic bilaterian must have had. Another, the number of cells in arthropods, is cited as an unpublished ms by Valentine. However, almost all of the counts boil down to one source, a critical source I haven’t yet been able to find. This very important paper, that purports to give cell type numbers for echinoderms, cephalopods, fish, amphibians, lizards, and birds, is:
Sneath, PHA (1964) Comparative biochemical genetics in bacterial taxonomy. pp 565-583 in CA Leone, ed. _Taxonomic biochemistry and serology_. Ronald, New York.
It’s a paper about bacterial taxonomy? And biochemistry? The only discussion in the text of the Valentine paper about this source mentions that it compares DNA content to cell type number, a measure that Raff and Kaufman have shown most emphatically to be invalid. And it’s from 1964, although the author seems to still be around and active in bacterial taxonomy and molecular biology right up until at least a few years ago. He doesn’t look like a histologist or comparative zoologist though, that’s for sure.
It’s from 1964. Oh, boy. I did manage to track down a copy of this volume in a library a few miles away, but I haven’t yet been able to get out and read it. I’m not too inclined to even try right now, because this appendix also has a little subscript in fine print at the bottom…virtually every source in this list, including Sneath, is marked with an asterisk, and the fine print tells us that that means “estimates NOT [my emphasis] documented by lists of cell types or by references to published histological descriptions”. In other words, there ain’t no data there, either.
I’m afraid to look up Sneath, for fear that it will turn out to be an estimate of cell number derived from measures of DNA content, with a bit of subjective eyeballing tossed in. At least that would explain why Kauffmann could find a correlation between DNA content and complexity, though.
From my perspective right now, this whole issue of cell type number is looking like a snipe hunt, a biological myth that is receding away as I pursue it. Does anybody know any different?
I didn’t have quick access to the all-important Sneath paper, but Mel Turner did, and he summarized it for everyone.
…there’s no original data. Here’s the relevant text:
“Although there are many possible correlations, for example, that between cell size and DNA content (135), it seems plausible to suggest that the amount of DNA is largely determined by the amount of genetic information that is required and that this will be greater in the more complex organisms. Fig. 38-2 shows the distribution of DNA contents of haploid nuclei taken from the literature, mostly from several compendia (4,10,87,128,134,135). The haploid nucleus was chosen for uniformity, and because the genetic information in diploids is presumably mostly reduplicated. The values are plotted against the number of histologically distinguishable cell types in the life cycle of the organism (suggested by a figure of Zimmerman (141)). This number is some measure of complexity, and was estimated from standard textbooks (5,13,85,126). In Fig. 38-2 organisms incapable of independent multiplication (e.g., viruses) have been assigned to the 0.1 cell level. The values for some well-known organisms are shown in Fig. 38-3.”
Fig. 38-2 is a graph of number of cell types (Y-axis) vs. log content of DNA/gamete, with a extra superimposed x-axis of “number of bits” (“one nucleotide pair = two bits”).No species names are indicated, but there are clusters of multiple separate points plotted for “mammals”, “birds”, “fish”, “angiosperms”, “bacteria” “algae & fungi”, “viruses”, etc. [oddly, he scores “RNA viruses” as having DNA content].
Fig. 38-3 purports to show “the histological complexity of some well-known organisms” with a log graph placing examples like “Man, Mammals” at the top with ca. 200 cell types, and “birds”, “reptiles”, “amphibia”, “fish” [again, no species names] just below that, then various cited generic names of plants animals, protists and bacteria [e.g., Pteromyzon (sic), Sepia, Helix, Ranunculus, Polypodium, Escherichia, etc.; about 50 taxa altogether]. Strictly unicellular organisms with different cell types during the life cycle [cysts, spores, gametes, etc. are properly scored as having histological complexity; e.g., Plasmodium scored with ca. 6 cell types]
There’s also discussion of the significance of the reported rough correlation of complexity and DNA content, a suggestion that histologically complex organisms should require disprortionately many times the DNA amounts of simple ones [cell specialization and regulation], a mention of some plants and amphibia with ‘unexplained’ very large DNA contents, and a page of stuff on base-pair changes, informational “bits”, & Kimura.
Table 38-3 “estimated amount of genetic and phenetic change in vertebrate evolution” looks pretty odd indeed [especially in a paper on bacterial biochemistry!]; it apparently tries to say something about times of origin and amounts of DNA change [% and in “bits”] for classes, orders, families, genera, species…. a bit dubious, to put it mildly.
Looking at the References list for the anatomical data sources cited for Figs 38-2 and 38-3, the “standard textbooks” were indeed just that:
5. Andrew, W. 1959. Textbook of Comparative histology. Oxford Univ. Press, London
13. Borradaile, L.A., L.E.S. Eastham, F.A. Potts, & J. T. Saunders. 1941. The Invertebrata: A manual for the use of students. 2nd ed. Cambridge Univ. Press, Cambridge.
85. Maximow, A.A. & W. Bloom. 1940. A textbook of histology. W. B. Saunders Co., Philadelphia.
126. Strasburger, E., L. Jost, H. Schenck, & G. Karsten. 1912. A textbook of botany. 4th English ed. Maximillian & Co. Ltd. London.
The Zimmerman citation from above is: Zimmerman, W. 1953. Evolution: Die Geschichte ihrer Probleme und Erkenntnisse. Alber, Freiburg & Munchen 623 pp.
Stephen Jay Gould wrote about a similar issue in Bully for Brontosaurus, in his essay on “The Case of the Creeping Fox Terrier Clone”, which describes how certain conventions, like describing the size of a horse ancestor as being as large as a fox terrier, get canonized in the literature and then get reiterated over and over again in multiple editions of textbooks.
This one isn’t as much a textbook problem as it is a deeply imbedded myth in the scientific literature. We haven’t even defined what a cell type is, yet somehow, again and again, we find papers and books claiming that it has been accurately quantified, and further, that it supports a claim of increasing complexity that puts humans at the pinnacle.
STOP IT.
I seem to have written about this problem every 6 or 7 years, to no avail. I’ll probably complain again in 2020, so look for a version of this post again, then.


“The lazy meme”
It is backed like a fox terrier…
The whole thing is garbage. In the organ of Corti, just one tiny part of the inner ear, you can identify 11 distinct cell types on the basis of distinct gene expression, which is as good a definition of a distinct cell population as any. There are at least 20 different types of retinal ganglion cells in the eye. So we’re up to 31 cell types in two sub-populations of two sensory organs alone. Pthhhhht.
Once again, an excellent post. I came across that graph in an evolution textbook that I used to use in my evo class (not anymore, btw). I confess I had not thought about the problem, only accepting its claim at face value. I now looked at that text figure, and found that the authors chose to substantially stretch the Y axis to give a stronger impression that the slope is steeper. Caveat emptor!
Also, millions of years ago seems a little off to me. Sure, Hominidae originated only recently, and Arthropoda go back past 500 million. But that’s not really comparing apples to apples; chordates are just as old, and small groups of arthropods didn’t stop appearing. Ctenophora would split off somewhere between Porifera, Cnidaria, and Bilateria, but their last common ancestor is only some 60-70 million years old.
Is it wrong to think this is also a problem here, or would it only amount to shading in the area below the chart were it not for the vertical axis being broken?
Why must we be more complex than flies? Their very name says that they fly. That can’t be easy. They have antennae, and more legs with more joints, and more eyes, and bodies that can keep functioning even without the head, and they do all that in a size we can’t work anything better than fingernails in. The little buggers must be tricky as hell inside.
I can understand wanting to measure the number of cell types and expecting some form of progression. If someone devised an objective, consistent standard for determining cell types, I think it’d be interesting to survey the animal kingdom to see how everything compares. It’d be fascinating to see how low some animals can go as well as find “simple” animals that go to ridiculous heights. I don’t expect humans to be at the top. It wouldn’t surprise me too much if we weren’t in the top 10%.
This myth is just pathetic. I do think that humans are an amazing species, but I wouldn’t fudge the numbers to pretend there’s an objective basis for that opinion or try to boil it down to a single measurement. I don’t always judge competing designs on the basis of which one has the most X. For some things, it’s better to sacrifice a little X to gain a lot of Y and Z.
It seems that every field has these fox terrier ideas that keep being reported. In my own field, software development, there are several such undocumented ideas, e.g. that a good programmer is 10 times as efficient as a bad programmer. Luckily, in software development, someone has started to look at these ideas, and have tracked down their origins, and found that there is nothing to them – he has written a nice little book called The Leprechauns of Software Engineering.
The Great Chain of Being lives! How many cell-types do angels have, I wonder? Do seraphim have more than cherubim?
The number if cell types must be the same as the number of bones in the human body – more evidence for fine tuning! (Maybe not)
Hey, 210 is my area code, so it is obviously a cool number.
I share your contempt of (just ran into this word somewhere and love it) assfax used in this way.
Not exactly addressing the question directly, but size of genome could also be used as a predictor of complexity, no? Problem is, we are way behind Euglena, which has a genome maybe three times bigger than ours. Makes sense if you’re half plant/half animal. I also agree with Andy Groves further up this post, there are not only at least 20 types of ganglion cell in the retina, but also at least 35 types of amacrine cells, 11 types of bipolar cells, … The cell type issue seems more related to greater knowledge of more commonly studied organisms – how many in-depth studies are there of sea slug anatomy (my apologies to sea slug experts).
There is, of course, an obvious reason that humans have more identified “cell types” than any other organism: We put much, much more effort into studying human cells than we do the cells of other species. Given that there is no fore-ordained typology, that means every specialization of functional or diagnostic importance gets recognized. Voila! We have lots of cell types.
There are a number of anatomy ontologies that catalog different cell types from a variety of organisms, in graph-like classification networks. These are frequently used in bioinformatics for purposes such as querying gene expression data.
These are not to be taken as indicators of numbers of “true cell types” whatever that might be – it’s always possible to refine a type into subtypes based on some kind of characteristic, and these ontologies reflect exactly the kind of biases mentioned by the aptly named photoreceptor, above.
However, just for fun, let’s look at some of the numbers:
The Drosophila (fruitfly) anatomy ontology has 3795 cell types listed – 1895 of which are types or neurons! (See the Virtual Fly Brain for a list, together with some lovely images and gene expression data).
The Zebrafish anatomy ontology has 577 cell types, whereas Xenopus anatomy (African clawed frog) ontology has 115.
The plant ontology has 195 cell types.
The porifera ontology (sponges), sitting at the base of the metazoans, has 46 cell types
The cephalopod ontology, lists a sad 7 <コ:{彡
The OBO cell ontology (covering a variety of species) has 2056 cell types – 555 of these are hematopoietic cells (this is certainly an underestimate of the number of groupings of cells coming from flow-cytometry experiments)
Again, these numbers do not necessarily correlate with organism complexity, cell type diversity or any other biological property. The numbers primarily reflect subjective classification choices, tractability of model systems for studying certain cell types, etc.
Either that or let me be the first to welcome our new Dipteran overlords..
Does anyone have some suggestions about appropriate Wikipedia articles and other places (does a biologist feel like writing a letter to the relevant journals, etc?) where this post should be linked, to at least *try* to get the word out about this BS myth? I mean, we aren’t going to be able to kill it all on our own, be we can dammed well make a dent…
I wonder how important “cell types” are anyway. How different is it to baraminology, with its obsessive focus on finding ways to combine as many different species as possible under the one “kind” in order to rationalise the Noah’s Ark story as a literal true history? What does number of cell types actually measure anyway? Complexity? I guess there’s probably a correlation. But what sort of complexity? Shannon? Kolmogorov? Krohn-Rhodes? State vector probability? Fractal? Constructal? And even if the correlation is causal, what does it mean? Is an organism with 200 cell types superior to an organism with 195 except in the trivial sense of ranking by number of cell types?
This post really should be put into the scientific literature somewhere. PZ, wouldn’t you like another easy publication for your CV? I’m thinking that TREE would be a nice fit. Or possibly Cell. What do you think?
A Wire Fox Terrier or Smooth Fox Terrier?
Just wonderful. A comment I make on Sandwalk leads to an entire post on shoddy thinking and myths in science. Great. Well, after explaining myself I might as well double-down.
I’m pretty sure I got the assertion about cell type number from S Kauffman. I remember thinking at the time that coming up with 210 cell types is ridiculous. On the other hand, it was reasonable to think that mammals have more cell types than flies without being precise. ( Kauffman needed precise numbers for an equation: fractals and complexity or somesuch?)
The whole discussion on SW was based on a post concerning the fact that vertebrates don’t have many more genes than most inverts. Does this mean that verts are minimally more complex than inverts or are there layers of regulatory complexity hidden in the junk? It seems to me that there must be objective criteria to suggest that verts are substantially more complex. Everything I’ve studied in biology invariably begins with: “here are the 3,4,5,10 homologs in vertebrates involved in process X and here is the 1 gene in Drosophila”. There are many more criteria one could consider and though there are caveats with all I cant help but to believe its not just another example of an inflated ego. And why it called the Inflated Ego Problem? Anyone looking to boost their ego wouldn’t be thrilled to find that mice, chickens and alligators, much less fish have about the same level of complexity as humans as judged by any of these criteria..
We all know that no organism is more ‘advanced’ than another and all living organisms are equally good at surviving, but most organisms have unique adaptations that allow them to survive. A certain level of morphological and behavioral complexity is the unique adaptation of vertebrates ( or at least amniotes) For the past 300 years science has been continuously knocking us off our pedestal and deflating our egos. However much we would like to think humans are special we find that we don’t have any truly unique traits. Just in my lifetime several of our defining characteristics have fallen by the wayside; tool use, self-awareness, empathy. But one can see this trend and extrapolate too far and conclude that we as mammals ( or verts) have nothing at all unique. I’ll call this the False Modesty Problem
What makes you say that? Certainly the examples with the most behavioral flexibility – in the sense of learned, reasoned, and cultural behaviors – are all mammals and birds. But the converse doesn’t work, since many other mammals and birds are easily exceeded by octopuses in those respects. And that’s selecting the one type of complexity that seems most favorable to us.
For instance, it assumes instinct is in some way simple, when it’s merely inflexible – look at for instance all the different behaviors of social insects like honeybees, slowly rotating their dances in the hives as the sun moves across the sky. And for morphological complexity, I don’t see it at all. The amniotes are impressive, to be sure, but nothing without rivals.
Mollusks and arthropods especially have their own complex organs, their own specialized parts, their own differentiated cells in complex assemblies. What criterion makes a squid with its muscle-controlled chromatophores, a spider with all its various silk and venom glands, or a mantis shrimp with its deadly appendages and incredible eyes is any inherently simpler than a lizard? The idea that complexity is a unique specialty of the vertebrates does not seem at all substantiated to me.
The cell type count of Diapsida that are all extinct hundreds of millions of years ago!? How did they determine that? Oh, right! BY PULLING NUMBERS OUT OF THEIR ASSES! Point in case.
My bad… My brain for some reason mistook Diapsida for Therapsida…
You want myths in biology? Pick up any introductory biology textbook and look up Mendel’s original experiments with garden pea plants. Look at the color illustration of the seven phenotypic characters Mendel supposedly tested. Is purple versus white flower color in the figure? Now, read Mendel’s paper describing the seven different characters in garden peas that he studied (you can find the original paper here, in the original German and in English translation: http://www.mendelweb.org/Mendel.html). Is purple versus white flower color in the list of characters tested? Interesting…
Toward the end of the paper, Mendel mentions that in a later set of experiments he tested flower color and found the same ratios that he found with the original seven traits he tested. According to the paper, he tested “violett-rothe und weiss Blüthenfarbe” (i.e. “violet-red and white blossom color”), but this test was NOT in his original set of seven experiments, which are the ones always illustrated in biology textbooks.
So, when did biology textbooks start this particular myth? As far as I can tell it was in the first biology textbook with full color illustrations: William T. Keeton’s Biological Science, 2nd ed. The illustrator thought he could kill two birds with one stone by illustrating the technique used to ensure controlled fertilization (i.e. removing the stamens from the flowers using iris scissors) and purple and white flower color. Except that Mendel didn’t study purple versus white flower color in his original series of seven crosses.
Which pair of traits did Mendel actually study, but were replaced by purple versus white flowers in all introductory biology textbooks? The color of the seed-coat, in which gray-brown is dominant and white is recessive. According to Mendel’s original paper, gray-brown seed coats are associated with (what we would now refer to as linked with) “violet-red blossoms and reddish spots in the leaf axils,” but once again Mendel did NOT explicitly test purple versus white flower color in the experiments for which he is remembered, and for which the science of “Mendelian genetics” is named.
And why is the clearly incorrect list (and colored figure) of the seven traits Mendel supposedly studied included in every introductory biology textbook today? Because Keeton’s textbook was the most widely used textbook in biology for decades, so all of the other publishers simply copied what was in his textbook as a way of gaining market share.
Can this myth be corrected now? How many professors’ sets of lecture notes and PowerPoint slides would have to be changed to correct this mistake, and how many textbooks would need new illustrations that included the correct list of the seven traits, and how many people would complain about these changes, or (even worse) suggest that Mendel really did study purple and white flower color in his original series of seven experiments?
Great reading, and good comments – props to science-minded commenters/scientists for collaborating to educate us laymen – and of course to PZ for taking time once in 6-7 years to correct another science myth.
This being said the number 210 cell types seems like a huge oversimplification for us humans – I find Chris Mungall’s 3795 number for Drosophila much more appropriate. Actual proper study with a common classification standard would actually be great – I want nice coloured diagrams with multiple complexety parameters taken into account for a host of species!
=8)-DX
Another interesting (and related) question is how many cells are average adult humans composed of? Carl Zimmer has a blog post that it’s about 37 trillion (I have been telling my students 100 trillion). Sounds like the right order of magnitude, but the number “37” bothers me. I recall reading somewhere that if you ask people to randomly choose a number between 1 and 100, most of them will choose either 37 or 47 (with 57 being third, as in Heinz’s varieties). Anybody know a good source for a reliable answer?
petrander:
Excuse the pedantry, but I have to point out that Therapsida isn’t extinct either, as you and all your friends are therapsids. They’re opisthokonts too.
Good comments people.
I wanted to call attention to Chris Mungall at #14 who has a squid emoticon. First I had seen that!
allanmacneil #23: You are right about the Mendel myth. That was a great point. I count myself among the instructors who are teaching it wrong, but the dang text books are hard to over-ride.
allenmacneill,
The ref is An estimation of the number of cells in the human body, Bianconi et al. Most are red blood cells.
The complexity of a system is the number of ways in which it can go wrong.
That’s my favourite definition, not because it is of practical use, but because it is the definition which made me think most about what we mean when we say something is complex. With this definition, perhaps humans come top of the complexity table, because there are so many types of mental disorder that we can suffer from. I’m not sure though. Fox terriers can be quite neurotic, for example. Perhaps we have bred more failure modes into dogs than we possess ourselves.
If there is a specific determined number (3795) of Sophophora melanogaster) cell types, then there should be a paper that identifies and ennumerates each cell type. If not, this number is bogous, i.e. each previously identified type cannot be identified by another researcher.
That “fox terrier” comparison was always vague to me since I’ve never been a member of the English fox-hunting tribe.
I have often thought that if metamorphosing parasites were writing the books, the obvious measure of complexity would be the number of different life-cycle stages that a species had developed–a true measure of evolutionary superiority.
rodw @19: It makes sense that an organism that undergoes metamorphosis, especially complete metamorphosis, has more cell types than organisms that remain in more or less the same shape all their lives. The original Sneath paper said that it looked at cell types throughout the life cycle but the paper using its numbers says that it ignored larval states. The whole thing is a mess that’s short on verified facts. Meanwhile, there are many ways to rationalize an assertion about complexity, depending on which group you favor. Invertebrates have a longer evolutionary history and thus have had more time to accumulate variations in cell type. Bony fish are among the most diverse vertebrates on Earth, while bats and birds are more derived than ground-dwelling mammals from the original body plan. Taxa that have undergone major changes in lifestyle, such as the cetaceans going from water to land to water again, may have had to develop new types of cells to cope with changes in the biochemical environment. The same goes for species that go from salt to fresh water and back. Man is a rationalizing animal.
Shame on you PZ. You’re ignoring the science of numerology, which is at least as relevant as any of the sources you mentioned. The number of cell types in a human is exactly the product of the first four prime numbers, the 187 cell types for a bird is 11×17, which uses two large nonconsecutive primes. Also, 187 = 14 squared – 3 squared, which is very important. The 50 cell types for a fly breaks down to 2x5x5, repeating a prime, which can’t be good.
All in all, this validates the methods used in all those papers you’re trying to dismiss.
Mothra – what do you mean by bogus? Yes, some of the classes in the ontology could be argued to be arbitrary groupings, I’m not making the claim that there are a specifically determined number of “true cell types”. The biologists and informaticians that make this resource have identified nearly 4,000 cell classes which are useful for annotating and analyzing gene expression, imaging data, etc.
You make a good point about references. In fact, many of the cell types in
the Sophophora (nice) ontology have references attached, as well as connections to other neuron types, location in the brain, etc. See for example:
http://www.virtualflybrain.org/site/tools/anatomy_finder/index.htm?id=FBbt:00003720
marcoli – glad you noticed the squid emoticon! I modified an existing one くコ:彡 and added a sad face (my squid was sad as he apparently had so few cell types)