Let me tell you about this miserable year I had in grad school. Judith Eisen and I had figured out that there was this repeating pattern of spinal motoneurons in zebrafish — this was special because it meant that we had a new set of identified neurons, cells that we could name and recognize and come back to in fish after fish, and that had specific locations and targets. I had flippantly suggested that we name them Primary Zebrafish Motoneurons (PZM cells, get it?), but a colleague, Walt Metcalfe, talked me down from that bit of vanity — it is so 19th century to name a cell after yourself, even indirectly — and I came up with the rather more mundane names of CaP, MiP, and RoP, for caudal, middle, and rostral primary motoneurons, for their location within each segment. So yay, interesting result, and it fit well within the overarching project I was working on for my thesis, which was on the development of connectivity in the spinal cord.
Specifically, I was looking at how another famously named neuron, the Mauthner cell, grew an axon down the length of the spinal cord and hooked up to the motor neurons there. Mauthner is a command neuron; when it fires, it sends a signal to one side of the spinal cord, triggering the motoneurons on that side to make all the muscles contract — the fish bends vigorously and quickly to one side as part of an escape response. Finding out that our one named cell, Mauthner, was making synapses on another set of named cells, our primary motoneurons, was an opportunity to look at connectivity in an even more detailed way.
But then my committee asked a really annoying question: how do you know Mauthner is making synapses on CaP? Have you looked? Thus began my miserable year. I said no, but how hard can it be? I’ll just make a few ultrathin sections, look at them in the transmission electron microscope, snap a few pictures, and presto, mission accomplished. Except, of course, I hadn’t done EM work before. Our EM tech, Eric Shabtach, made it look easy.
So I started learning how to fix and section zebrafish embryos for EM. It turns out that was non-trivial. I was working with nasty chemicals, cocktails of paraformaldehyde, glutaraldehyde, and acetaldehyde, which all had to be just right or you’d end up with tissue blown up full of holes. I had to postfix with osmium tetroxide, with all the fun warnings about how just the fumes can fix your corneas. And then I had to master using an ultramicrotome and making glass knives, and cutting those fish just right. There were times I’d get the fixation perfect and then find I’d screwed up on the sectioning, and produced a lot of crap as the knife chattered across the section, or there was a bit of a nick in the blade that gouged furrows across every one. And then the way we got these extremely thin slices into the scope was to scoop them up on these delicate copper grids, and of course every time you were closing in on the synapse you wanted, that section would have the most interesting part fall right on an opaque copper grid wire. Or you’d find that that was the section you lost.
It takes a lot of skill and practice to do electron microscopy well, and it also takes a little luck, at least in the old days, to find the one thing you were looking for. I failed. I struggled for about a year, going in every day and prepping samples and spending hours slicing away at tiny dead embryos imbedded in epoxy, before finally giving up and deciding I needed to do stuff that was more immediately successful, because I needed to do this graduation thing.
I still kind of cringe remembering that long fruitless year, but now I can ease my conscience by just telling myself the technology wasn’t yet ready. Here’s a cool new paper, Whole-Brain Serial-Section Electron Microscopy In Larval Zebrafish. They’ve automated the process. Just look at this goddamn machine, it’s beautiful:
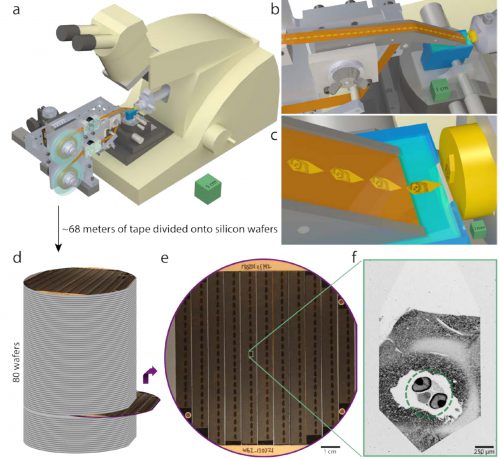
Serial sectioning and ultrathin section library assembly for a 5.5dpf larval zebrafish. a, Serial sections of resin-embedded samples were picked up with an automated tape-collecting ultramicrotome modified for compatibility with larger reels containing enough tape to accommodate tens of thousands of sections. b–c, Direct-to-tape sectioning resulted in consistent section spacing and orientation. Just as a section left the diamond knife (blue), it was caught by the tape. d, After serial sectioning, the tape was divided onto silicon wafers that functioned as a stage in a scanning electron microscope and formed an ultrathin section library. For a series containing all of a 5.5dpf larval zebrafish brain, ~68m of tape was divided onto 80 wafers (with ~227 sections per wafer). e, Wafer images were used as a coarse guide for targeting electron microscopic imaging. Fiducial markers (copper circles) further provided a reference for a perwafer coordinate system, enabling storage of the position associated with each section and, thus, multiple rounds of re-imaging at varying resolutions as needed. f, Low-resolution overview micrographs (758.8×758.8×60nm3 vx –1) were acquired for each section to ascertain sectioning reliability and determine the extents of the ultrathin section library. Scale boxes: a, 5×5×5cm3 ; b, 1×1×1cm3 ; c, 1×1×1mm3 . Scale bars: e, 1cm; f, 250µm.
Then they scanned in all those tidily organized thin sections into the computer for reconstruction. I am impressed.
We next selected sub-regions within this imaging volume to capture areas of interest at higher resolutions using multi-scale imaging. We first performed nearly isotropic EM imaging by setting lateral resolution to match section thickness over the anterior-most 16000 sections. All cells are labelled in ssEM, so this volume offers a dense picture of the fine anatomy across the anterior quarter of the larval zebrafish including brain, sensory organs (e.g., eyes, ears, and olfactory pits), and other tissues. Furthermore, this resolution of 56.4×56.4×60nm3/vx is ~500× greater than that afforded by diffraction-limited light microscopy. The imaged volume spanned 2.28×108 µm3 , consisted of 1.12×1012 voxels, and occupied 2.4 terabytes (TB). In this data, one can reliably identify cell nuclei and track large calibre myelinated axons. To further resolve densely packed neuronal structures, a third round of imaging at 18.8×18.8×60nm3/vx was performed to generate a high-resolution atlas specifically of the brain. The resulting image volume spanned 12546 sections, contained a volume of 5.49×107 µm3 , consisted of 2.36×1012 voxels, and occupied 4.9TB. Additional acquisition at higher magnifications was used to further inspect regions of interest, to resolve finer axons and dendrites, and to identify synaptic connections between neurons.
Thirty five years ago we were storing most of our image data on VHS tape, and our computers all used floppies with about 100K capacity. I wonder how many floppies we would have needed to store all that? Oh, I did get my very first hard drive about the time I graduated, which held five million bytes. I was very proud.
I was wondering if they actually had the EM section demonstrating the Mauthner-to-CaP synapse. Probably. Now it’s such a minor issue that has already been shown elsewhere and with multiple techniques that it isn’t even mentioned. It’s in their data set, though, I’m sure. They’ve reconstructed the entire axon arbor of CaP in serial EM sections.
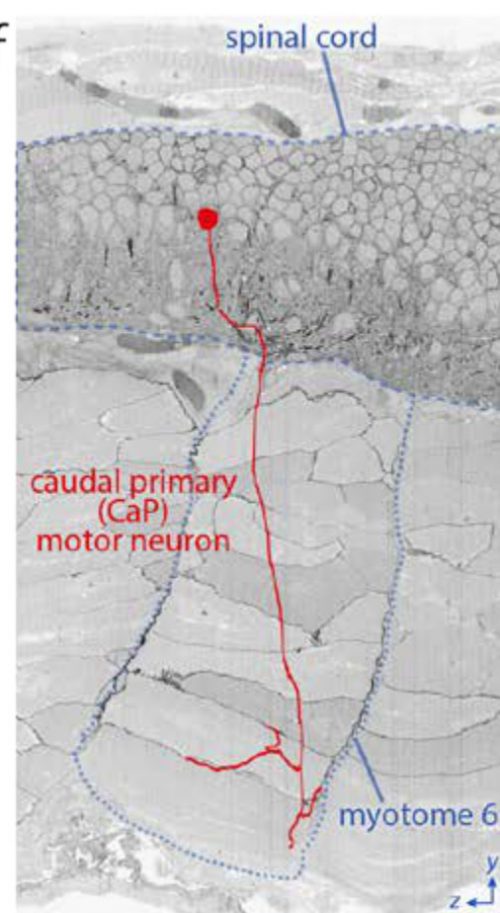
The position of a caudal primary (CaP) motor neuron in the spinal cord and its innervation of myotome 6 projected onto a reslice through ~2200 serial sections.
2200 sections! I spent a year on that project and probably got half that number. I don’t know whether to cry or steal the data, invent a time machine, and go back and hand myself a photo at the start of that year.


Electron microscopy? machines are nice, but you probably needed a few books to guide you as well.
Jesus is like my Scanning Electron Microscope: (a scientist looks at his relationship with the Creator)
by Mark Armitage
OsTet!! Yaayyy!!
And fuck me, that’s gorgeous!
But they’ve taken all the fun out! The first time I can remember losing all sense of time due to being completely immersed in lab work was cutting ultrathin sections. And no, not because of the ether we used to smooth out the wrinkles.
But seriously, that’s great.
But really, cutting ultrathins is fun, too.
@carlie I know, right? The bleeding lacerations from the Weck prep blades and glass knives! The partially fixed digits and the stink of aldehydes! The glowering and threatening of the PI because of too much “chatter!” The joy of being covered in vacuum pump oil! The certainty of leading a sad, lonely dead-end life! Ah, good times, good times.
Voxel is my new word of the day!
And …2.4TB from one run? That’s a lot of data to store, process, retrieve, transfer etc: new jobs in science will be tweaking and looking after these machines and never getting any spare time to science anything.
It’s a common thing in science: Think of how hard it was just a decade or two ago to sequence a genome. Or think of archaeologists fighting their way through a jungle in search of archaeological sites when today “we” can just use LIDAR and sometimes even discover sites adjacent to the earlier explorer’s path.
At least you did not have to look out for poisonous snakes or worry about tropical diseases. And if worst came to worst someone could have called 911.