Recently, Carl Zimmer made a criticism of the computer animations of molecular events (it’s the same criticism I made 8 years ago): they’re beautiful and they’re informative, but they leave out the critical aspect of stochastic behavior that is important in understanding the biochemistry. He’s talking specifically about kinesin, a transport protein which the animators are particularly fond of illustrating.
Every now and then, a tiny molecule loaded with fuel binds to one of the kinesin “feet.” It delivers a jolt of energy, causing that foot to leap off the molecular cable and flail wildly, pulling hard on the foot that’s still anchored. Eventually, the gyrating foot stumbles into contact again with the cable, locking on once more — and advancing the vesicle a tiny step forward. This updated movie offers a better way to picture our most intricate inner workings…. In the 2006 version, we can’t help seeing intention in the smooth movements of the molecules; it’s as if they’re trying to get from one place to another. In reality, however, the parts of our cells don’t operate with the precise movements of the springs and gears of a clock. They flail blindly in the crowd.
The illusion of directed, purposeful movement is a simplifying shortcut: as Zimmer describes, there actually is a lot of noise in the system, it’s just that the thermodynamics of the interactions promote a directionality to the motion. This is Chemistry 101. I figured that everyone with an undergraduate level of understanding of molecules would be able to grasp this.
I did not take into account willful ignorance, however. Jonathan Wells is angry that anyone dared to question the perfect “stately grace” of molecular machines, and accuses proponents of stochastic motion of Flailing Blindly: The Pseudoscience of Josh Rosenau and Carl Zimmer. He has a Ph.D. in biology, and he doesn’t understand what I just said was Chem 101? For shame.
But that’s not what the biological evidence shows. In fact, kinesin moves quickly, with precise movements, to get from one place to another. A kinesin molecule takes one 8-nanometer “step” along a microtubule for every high-energy ATP molecule it uses, and it uses about 80 ATPs per second. On the scale of a living cell, this movement is very fast. To visualize it on a macroscopic scale, imagine a microtubule as a one-lane road and the kinesin molecule as an automobile. The kinesin would be traveling over 200 miles per hour!
The speed of the reaction doesn’t say anything about the specifics of the molecular movement…and it’s especially not convincing when your trick is to multiply the actual speed in the cell by approximately 1012 to scale it up to the size of a car. The flow rate of the Mississippi river is about 1.5 miles per hour here in Minnesota — if you multiply that by 1012, oh my god, the water is moving at about 2000 times the speed of light!
But let’s set aside the stupid inflation for a minute. Wells cites a couple of papers to back up his claim of the rate of ATP consumption. It’s true. But it doesn’t show that the movement is steady and machine-like and precise at all. He must be trusting us to not bother even reading the paper.
Here’s the deal: we can actually watch single molecules of kinesin behaving. The typical trick is to use a fluorescent bead, attach that to kinesin, and then record the glowing bead’s movement as it is moving along with optical-trapping interferometry. That’s the problem with Wells’ accusation: we actually see the behavior, and it’s not linear, smooth, and graceful.
This is the data that the paper used to measure the quantum, jerky behavior of kinesin. Just look at the top graph: that’s a record of the bead’s movement over time. You should be able to see that the line holds steady at one distance for variable lengths of time, and then jerks upward. The “jerks” are distances of about 8nm, and the other graphs are power spectra to show that there is a peak periodicity of 8nm. It shows the opposite of what Wells claims; there are long pauses and sudden shifts in the directly observed track of kinesin movement. The 8nm emerges because when one “foot” of kinesin releases and wobbles forward to connect to tubulin, it has an 8nm step.
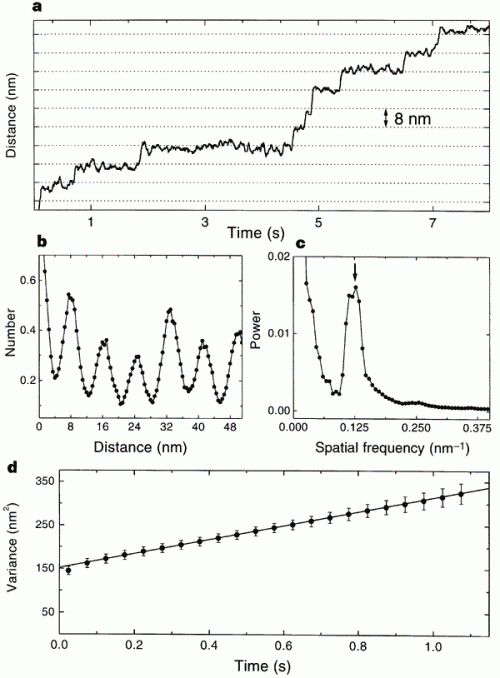
Horizontal grid lines (dotted lines) are spaced 8 nm apart. Data were median-filtered with a window width of 60 ms. b, Normalized histogram of pairwise distances between all pairs of data points in this record, showing a clear 8-nm periodicity. c, Normalized power spectrum of the data in b, displaying a single prominent peak at the reciprocal of 8 nm (arrow). d, Variance in position, averaged over 28 runs at 2 microM ATP (dots), and line fit over the interval 3.5 ms to 1.1 s. The y-intercept of the fit is determined by equipartition, left fencex2right fence = kT/alpha, where alpha is the combined stiffness of the optical trap and bead–microtubule linkage. The rapid rise in variance at short times reflects the brownian correlation time for bead position.
You can find lots of papers with direct observations of kinesin movement. Here’s data from another paper that essentially shows that the two ‘feet’ of kinesin alternate in their movement, because they made recombinant, asymmetric kinesin with slightly different step distances. Again, note the long dwell times punctuated with surges of movement.
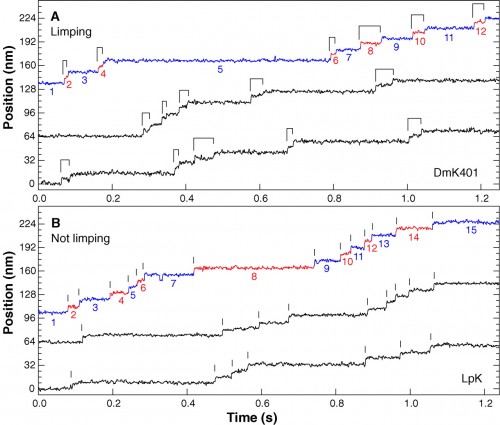
Representative high-resolution stepping records of position against time, showing the single-molecule behavior of kinesin motors under constant 4 pN rearward loads. (A) Limping motion of the recombinant kinesin construct, DmK401. The dwell intervals between successive 8-nm steps alternate between slow and fast phases, causing steps to appear in pairs, as indicated by the ligatures. (B) Nonlimping motion of native squid kinesin, LpK. No alternation of steps is apparent; vertical lines mark the stepping transitions. Slow and fast phase assignments, as described in the text, are indicated in color on the uppermost trace of each panel (blue and red, respectively), and the corresponding dwell intervals are numbered. All traces were median filtered with a 2.5-ms window.
The contrast is between “stately grace” and “jiggle and jump”. The evidence shows that it is the latter. Yet, somehow, Wells closes his weird series of non sequiturs with this question, as if he expects everyone to give him the answer he wants.
So, who are the pseudoscientists?
The answer is obvious. Wells and his cronies at the DI.
Asbury CL, Fehr AN, Block SM (2003) Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302(5653):2130-4
Schnitzer MJ, Block SM (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature388(6640):386-90.
By the way, I found about this because the Discovery Institute sent me email (silly season and all) proudly announcing that they were about to release evidence that would clearly rebut Zimmer’s assertion that kinesin “flails blindly” — they are making a computer animation showing that it doesn’t.
Yes, they are that stupid.


Nice.
I’m worried now that we’ll have to have our comments peer reviewed. :-)
Thanks for this! I appreciate the glimpse into biochemistry–after ending my own chemistry studies with a gentleman’s B in second-semester inorganic almost 30 years ago.
The kinesin video captivated me a year or so ago, and made my think I might have kept going if I knew the neat stuff that lay in store. I wondered about proteins managing to behave as such clockwork machines, and this post of yours answers it comprehensibly. Your graphs look like a great illustration of a one-dimensional random walk with a very slight directional bias.
Is there a total lay-person’s explanation why the net effect is directional?
This manuscript proposes an interesting hypothesis that should be published, but unfortunately it entirely fails to test it. I recommend publication after substantial revision.
Going forward is slightly easier than going backward; it releases more energy (as heat).
Ah, but, those graphs and other sciencey-smelling things are watching the beads, which are interfering with the natural grace of the
evolveddesigned mechanism. The leggy-things aren’t meant to be hauling around a ball-and-chain, no wonder they flail!Take that, athesit evilutionistas!
I hates over-simplified animation.
Last time this came up, with impossible machines in cells and all, David M. said it was all just electrostatics and Brownian motion.
David Marjanović
But that was testing it…
That data is beautiful to look at! Much more beautiful than the silly animations.
You must be new here, because THIS. IS. PHARYNGULA!*
*where all claims of natural truth are dissected mercilessly until the only thing left is the bare naked germ of supportable evidence**
**Thank you, and have a nice day. :)
I suppose I could go look up the paper, but asking here is easier:
I was somehow under the impression that only the rear foot can bind ATP and lift up ( steric hindrance or somesort) The loose foot can then take a step forward or bind back down to the same place, wasting an ATP. Is this correct? If so wouldnt it be more like an average of 1.5 ATPs per step?
Citation needed.
Thanks for this, PZ. Great take-down. I have alerted a few other people to the post.
Thanks David M. for the explanation about moving forward being slightly easier than going backward.
All in all, a good day for learning something new.
Wait – Jonathan Wells didn’t throw himself on his Dear Leader’s funeral pyre?
:-(
I watched the new video. The molecules jitter as if the video was a very amateurish stop motion film. That’s a good thing, because I was wondering how they’d convey the randomness. I think it does an excellent job. The processes may sound elegant when you describe them like our middle world machines, but one of the lessons I’ve learned from listening to my chemistry teacher and people who know something about quantum mechanics is that the world of the tiny is much messier than we’re used to thinking about. We’re not used to seeing all the little bumps and missteps, just the big net effect.
I took AP (advanced placement) Chemistry in high school, and Ms. Isbell was a good enough teacher that I was able to CLEP out of 6 hours of college chemistry years later with only a little cramming on the math (which wasn’t as big an issue as I thought it’d be). One of the things she managed to keep me aware of was equilibrium. Things don’t necessarily sit still just because you’re not doing anything. If you’ve got a salt solution with a crystal of that salt, some of the molecules in solution will be joining the crystal while others break off to join the solution. It’s a continuous process. It’s a matter of available energy and entropy that decides whether the system as a whole averages in one direction or the other. Left alone, the system will eventually reach a point where the two reactions happen at the same rate, balancing each other. The only way you can stop it is perfectly crystallize everything at 0K. (Good luck with that.)
I’m no biologist, but to me, life is funky chemistry because it’s excessively complicated, and there’s all sorts of complex chemicals being produced just to keep the whole mess from settling down into ‘tamer’ chemical reactions. Life is a messy, dynamic balance with conflicting chemical forces pulling us in a million directions at once. That’s why life is so delicate. Pharmacology is about carefully pulling the equilibria in a favorable direction. Toxicology is about pulling too hard in any direction. All the moving parts being continuously replaced and repaired means that sooner or later, something is going to break and send the organism crashing down or puttering to a halt.
That’s one reason I think life isn’t designed. Designed objects tend toward simplicity and direction because they have a purpose. Life as we know it is a messy chemical process that makes its own purpose after it’s already here.
It doesn’t help that many living organisms have to survive by disrupting the delicate balance of other living organisms who have their own purposes in mind. The rabbit wants to use its body for a certain purpose that ends up making more rabbits. The wolf wants to use the rabbit’s body as a source of chemical energy. Why design two things that want to use the same resource for two conflicting purposes and determine the winner with trial by combat?
If you want to directly watch the motion of a similar ‘molecular machine’, myosin moving on actin, a visualization using high speed atomic force microscopy is
here.
The topic of myosin directionality is an area of active research. I’m not an expert, but I was impressed by this paper by Warshel published last year that used computer simulations to explain directionality. The paper was even described in Warshel’s Nobel Prize acceptance speech. Though, just today it was challenged in a letter to the Proceedings of the Natural Academy of Sciences, claiming the model is insufficient. They claim the complete picture is that:
Sorry for the academic paywalls!
News flash to Wells: *all* molecules move that fast. Because they’re really small, get it?
I remember learning about kinesin from Shane Killian’s How Evolution is Scientific series. Oh, for the days before he descended into crazed, homophobic libertarianism.
Here is a video made back in the hairy days of 1971 that manages to convey the stochasticity of molecular motion very cleverly — without any animation. The scientific content is somewhat out of date but not the part showing how stochastic things are.
Gaaahh! Wrong _and_ annoying in one sentence.
It’s not a high energy molecule*, it’s a molecule with a labile bond that has conveniently low enough energy as to be useful for enzymes to be able to do stuff with it.
Think about bribing a schoolful of kids to clean your car. 20c pieces are going to be a lot more useful than 500€ notes. And no, the fact that half the textbooks (that I used, maybe they’ve improved) use the term, and Wikipedia, and most of the internet, doesn’t make it right. Just more annoying.
*What does that mean anyway? Is it moving particularly fast?
@18 Joseph Felsenstein . That was smashing – thanks!
Those were VERY hairy days.
#13 Bronze Dog
Your comments about the salt solution really can be extended to the entire universe. The universe is constantly giggling (and yes I mean small laugh) at the atomic and subatomic levels isn’t it:-)
Joseph @#18 – that is an awesome video. I love “Initiation Factor Two”s outfit.
I certainly know that my understanding of chemistry improved by leaps and bounds once I grokked the concepts of dynamic equilibrium and directional tendency in otherwise disordered movement. It’s a very important point to get, so it’s rather sad if a phd doesn’t understand it. How does he manage to do his job?
The other thing I find a bit annoying about these videos, although i understand it is done for purposes of illustration so that one can see the points of interest, is that water is treated like an indivisible substance, appearing as it does on the scale of normal human experience. Never mind all the other molecules constantly bumping about. These CGI animations should all come with a few notes explaining how they are model illustrations.
Kinesin walking:
HHMI did decently good work a few years ago trying to capture the speed and random movements that go along with molecules. They have a series on DNA synthesis/transcription/translation with nice animations. Here’s the one on translation.
Something that none of these animations show is that at this scale, the water the molecules are floating would be visibly a bunch of Mickey Mouse head-shaped blobs. Yes, I know they have to delete the water molecules to show the interesting stuff, but they could start the whole thing off showing the water then transparency them out.
It would help eliminate the misconception that these are little machines. At this scale, even water is a bunch of standardized lumps.
@25 voss
Wells also screwed up by not mentioning the little popping noises made by the release of phosphate and the whooshing made when the trailing head moves forward. What kind of “intelligent” design would fail to include a full multimedia experience for our benefit? Next he’ll be telling us angels are silent in a vacuum…
I tell you, it’s enough to make me think HE wasn’t designed correctly.
Depending on level of magnification, there ought to be a few cytoskeletal elements getting in the way too, but same problem.
For an amazingly good discussion of how these kinds of molecular motors work check out the incredibly well written book: Life’s Ratchet. It explains the physics and biochemistry clearly and in excellent detail. The trick is that it takes energy to erase information. These molecules are as likely to “walk” forwards as backwards, but a blast at ATP at the right time erases the information needed for a backwards step. (It’s not quite that simple, but that’s the general idea.) Otherwise, movement is a random process powered by random interactions with ATP which is turned into ADP, releasing enough energy that, at macro levels, the reaction would take place at 5,000-10,000C.
P.S. For some myosin “walking” movies from an atomic force microscope, check out http://www.nature.com/nature/journal/v468/n7320/full/nature09450.html
This really bothered me about my second to last biology class I took. It was in high school, so it wasn’t even undergrad college level 101 yet, but we learned about some “simple” biological processes. (I got to transcribe, ironically, a lengthy professional article on protein synthesis into layman’s terms!) But I asked the teacher how does blah-blah this tRNA know how to attach to the ribosome here? Like, what was directing it? I had no understanding of stochastic behavior, and what bothered me was both the fact that the teacher had no understanding either, but more importantly that the teacher didn’t just say, “I don’t know.” They just said, “It knows how to get there.” I regret to say I abandoned biology when getting accepted into university and went into computer science instead. Probably would have been happier if I had stuck with biology. It’s far more interesting to me.