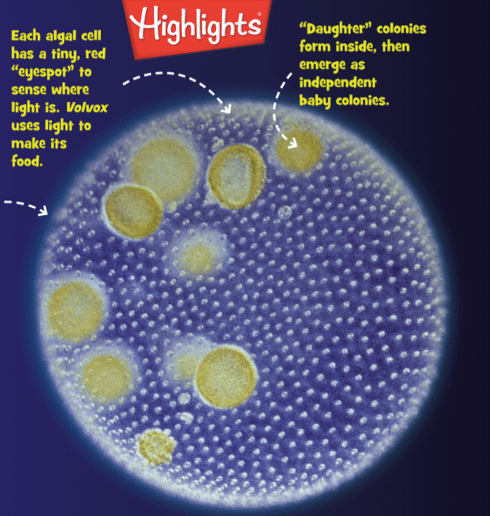
This is so cool! The March issue of Highlights Magazine (“Fun with a purpose”) includes a full page on Volvox. Highlights is aimed at kids 6-12 and publishes both print and digital editions:
In every 40-page issue, kids explore new topics, investigate cool subjects and find out about the world. Highlights magazine for kids is filled with stories, games, puzzles, riddles, science experiments, craft projects and interactive entertainment!